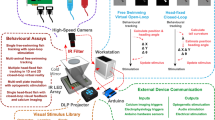
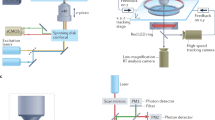
Abstract
A long-standing objective in neuroscience has been to image distributed neuronal activity in freely behaving animals. Here we introduce NeuBtracker, a tracking microscope for simultaneous imaging of neuronal activity and behavior of freely swimming fluorescent reporter fish. We showcase the value of NeuBtracker for screening neurostimulants with respect to their combined neuronal and behavioral effects and for determining spontaneous and stimulus-induced spatiotemporal patterns of neuronal activation during naturalistic behavior.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kim, C.K. et al. Nat. Methods 13, 325–328 (2016).
Looger, L.L. & Griesbeck, O. Curr. Opin. Neurobiol. 22, 18–23 (2012).
Ahrens, M.B. & Engert, F. Curr. Opin. Neurobiol. 32, 78–86 (2015).
Portugues, R., Feierstein, C.E., Engert, F. & Orger, M.B. Neuron 81, 1328–1343 (2014).
Vladimirov, N. et al. Nat. Methods 11, 883–884 (2014).
Ahrens, M.B. et al. Nature 485, 471–477 (2012).
Muto, A., Ohkura, M., Abe, G., Nakai, J. & Kawakami, K. Curr. Biol. 23, 307–311 (2013).
Naumann, E.A., Kampff, A.R., Prober, D.A., Schier, A.F. & Engert, F. Nat. Neurosci. 13, 513–520 (2010).
Kalueff, A.V. et al. Zebrafish 10, 70–86 (2013).
Orger, M.B. & de Polavieja, G.G. Annu. Rev. Neurosci. 40, 125–147 (2017).
Bruni, G. et al. Nat. Chem. Biol. 12, 559–566 (2016).
Nguyen, J.P. et al. Proc. Natl. Acad. Sci. USA 113, E1074–E1081 (2016).
Grover, D., Katsuki, T. & Greenspan, R.J. Nat. Methods 13, 569–572 (2016).
Haesemeyer, M., Robson, D.N.N., Li, J.M.M., Schier, A.F.F. & Engert, F. Cell Syst. 1, 338–348 (2015).
Guggiana-Nilo, D.A. & Engert, F. Front. Behav. Neurosci. 10, 160 (2016).
Hussain, A. et al. Proc. Natl. Acad. Sci. USA 110, 19579–19584 (2013).
Randlett, O. et al. Nat. Methods 12, 1039–1046 (2015).
Prevedel, R. et al. Nat. Methods 11, 727–730 (2014).
Mertz, J. Nat. Methods 8, 811–819 (2011).
Grosenick, L., Marshel, J.H. & Deisseroth, K. Neuron 86, 106–139 (2015).
Chen, X. & Engert, F. Front. Syst. Neurosci. 8, 39 (2014).
Shcherbakov, D. et al. PLoS One 8, e64429 (2013).
Pertuz, S., Puig, D. & Garcia, M.A. Pattern Recognit. 46, 1415–1432 (2013).
Kimmel, C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B. & Schilling, T.F. Dev. Dyn. 203, 253–310 (1995).
Takeuchi, M. et al. Dev. Biol. 397, 1–17 (2015).
Knogler, L.D., Markov, D.A., Dragomir, E.I., Štih, V. & Portugues, R. Curr. Biol. 27, 1288–1302 (2017).
DeMaria, S. et al. J. Neurosci. 33, 15235–15247 (2013).
Pardo-Martin, C. et al. Nat. Methods 7, 634–636 (2010).
Li, X. et al. PLoS One 7, e40508 (2012).
Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H.K. Science 322, 1065–1069 (2008).
Fahrbach, F.O., Voigt, F.F., Schmid, B., Helmchen, F. & Huisken, J. Opt. Express 21, 21010–21026 (2013).
Acknowledgements
We thank K. Kawakami and A. Muto (National Institute of Genetics (Japan)) for sharing tg(gSA2AzGFF49A;UAS:GCaMP7a) and tg(gSA2AzGFF152B), here referred to as GR152:Gal4, which was provided to us by R. Portugues and L. Knogler (Max Planck Institute of Neurobiology), K. Asakawa (National Institute of Genetics (Japan)) for providing tg(mnGFF7;UAS:eGFP), J. Ngai (University of California, Berkeley) for tg(OMP4; UAS:GCaMP1.6), M. Ahrens (Janelia Research Campus) for sharing tg(HuC:H2B-GCaMP6s), H. Baier and K. Slanchev (Max Planck Institute of Neurobiology) for sharing tg(HuC:Gal4;UAS:eGFP) and tg(HuC:Gal4;UAS:GCaMP6s), and L. Godinho (Technical University of Munich) for providing tg(HuC:Gal4;UAS:GCaMP6s). We are grateful to J. Huisken and F. Fahrbach for providing assistance on building the SPIM; J. Fuchs assisted with the SPIM setup and experiment. We thank B. Wolfrum, P. Rinklin, J. Rebling, D. Razansky and the central machine shop of Helmholtz Zentrum Muenchen for help with the design and production of the arenas; H. Rolbieski for administrative support and help with data organization; T. Durovic and J. Ninkovic for assistance with the two-photon microscopy experiments; M. Fabiszak, C. Penningroth and L. Garrett for valuable comments on the manuscript. We are grateful for support from the European Research Council under grant agreements ERC-StG: 311552 (G.G.W., A.L., P.S.) and the Helmholtz Alliance ICEMED (G.G.W.).
Author information
Authors and Affiliations
Contributions
P.S. designed and implemented NeuBtracker with important contributions from H.J., M.K. and A.C.; programmed control software; conducted neurobehavioral imaging experiments including SPIM; analyzed data; and wrote the manuscript. A.L. designed and implemented neurobehavioral and two-photon experiments, conducted p-ERK imaging experiments, analyzed data, created illustrations and wrote the manuscript. A.S. and T.L. implemented image registration routines and improved tracking algorithm and analyzed data together with S.S. M.C. carried out the first implementation of tracking algorithm and performed original image and signal processing. C.C.P. carried out the initial implementation of SPIM. A.M. conducted photoaversion experiments on immobilized fish. A.S. was responsible for animal husbandry and provided support for neurobehavioral imaging. S.R. generated tg(HuC:GCaMP6s). W.W. and V.N. supported the project and provided feedback on the manuscript. G.G.W. conceived and supervised the work, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Overview of NeuBtracker in MacroZoom configuration.
(a) Photograph of the system in its MacroZoom configuration (b) Zoom-in on the core system with labeling of the core parts; galvanometric mirrors (Galvo xy) for tracking, the electrically focus-tunable lens (ETL) for focusing, as well as the objectives used for excitation and detection by the two imaging channels (Beam splitter (BS) and dichroic mirror (DC)) (c) Drawing of the main components and the optical path to supplement the schematic in Figure 1. The inset on the upper right shows galvanometric scanning of a transparent composite resolution chart (with a fluorescent plastic slide placed underneath, 1951-USAF part: 3.7 x 3.7 mm) via the 15-fold magnified fluorescent channel. Individual tiles were acquired with overlap to allow for image stitching as shown below. The image on the lower right shows cellular features resolvable in a reporter line with sparse labeling of neurons throughout the anterior-posterior axis tg(OMP4; GCaMP1.6 x mnGFF7; UAS:eGFP). The scale bars represent 100 μm. All parts of NeuBtracker are compiled in a parts list that can be found in Supplementary Table 1 and together with 3D CAD assemblies on neubtracker.org.
Supplementary Figure 2 Overview of the modular configurations of NeuBtracker as an imaging and interrogation platform.
(a) Schematic of the combinations of the components used in the two configurations reported in this manuscript (‘MacroZoom’ and ‘MicroFixed’) and possible add-ons. The insets (upper left corner) gives a photographic overview of the alternative MicroFixed configuration which uses a microscope objective and fixed magnification. Several additional features were also tested such as coupling of additional lasers (either illuminating the whole arena or providing localized photostimulation via the galvanometric mirrors), or the use of motorized stages for high-throughput screening applications. (b-d) Different types of image contrasts that can be obtained by alternative configurations of NeuBtracker. As an example, the IR-illumination ring can either be positioned on top of the sample (IR-EPI inlet in (b)) or underneath it (IR-TRANS) to achieve epi- or transillumination. Furthermore, the addition of extra optical elements to the path can be achieved conveniently. A diffuser can be added for a projector to be placed under the sample (c- upper right). In this case, the noise level of the fluorescence channel is increased due to the bleed-through of backscattered excitation photons ((d)-middle right, images from MicroFixed configuration). By using a rotating holder, a dichroic can be positioned to deflect the excitation light after it passes through the sample. This results in images such as shown in c -top right. The presence of a sliding holder (or potentially a filter-wheel), enabled us to create the overlay in (bottom of c) were the same sensor captured fluorescence (green overlay) and directly afterwards, a brightfield image (gray) using an autofluorescent plastic plate as a 510 nm light source to create transillumination contrast utilizing the high dynamic range of the camera sensor. (d) To showcase the features that can be differentiated in the 7x magnified fluorescence channel of NeuBtracker MicroFixed, we show how the 7x channel may also be used to obtain anatomical details in reflection images with fast temporal sampling (5 ms exposure time) when an autofluorescent plastic slide (Chroma) is placed underneath the arena. The three frames on the right show movements of the fins and the heart displayed on a difference map (red for positive, blue for negative intensities with respect to the previous frame)
Supplementary Figure 3 Calibration of NeuBtracker components.
(a-c) A composite resolution target (a) was placed on top of an autofluorescent plastic slide and scanned on the MicroFixed configuration by translating the FOV of the 7x fluorescent channel to generate (b) a stitched image of the entire resolution target at high resolution. Individual tiles are shown in (c) (d-f) Both positive and negative resolution targets consisting of concentric rings are placed in the sample holder to ensure proper registration of the FOVs of the two cameras. (g-i) Profile of the fluorescence excitation analyzed by using an autofluorescent plastic slide. The red circle in (h) indicates the diameter of the arena. (i) Corresponding plot of the standard deviation/mean for all xy-galvo positions to quantify the homogeneity throughout the arena (indicated by the red area).
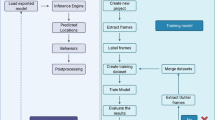
Supplementary Figure 4 Image acquisition automation and data processing pipeline.
Image acquisition automation and data processing pipeline (a) Simplified schematic showing the main connections to and from the console running the control software. (b) Screenshot acquired during one of the acquisitions, showing the GUI of the control software displaying both of the imaging channels (left: 1x behavioral tracking, right: magnified real-time view of the fluorescent imaging). (c) Flowchart of the data acquisition and processing pipeline with (d) examples from the feature matching of the SURF registration. Scalebars represents 1 mm.
Supplementary Figure 5 Synchronization of hardware components.
(a) In addition to the timestamps obtained by the computer clock, we have recorded signals directly from the components of the system using different types of output (galvanometric mirrors: analog voltage, fluorescence camera: TTL pulses, 1x IR camera: PLC logic requiring additional input voltage). (b) Example raw signals from a 50 μs time window during which the 1x IR camera acquired at 200 Hz and magnified fluorescence images were obtained at 50 Hz (the beginning of the respective exposure times correspond to the high values in the red/green lines respectively). The blue and cyan signals are the control voltages to the galvanometric mirrors. (c) Image of the resolution target acquired for illustration by the 1x camera. (d) Example signals from the galvanometric mirrors from a raster scan of the resolution target shown in (c) resulting in 12 consecutive images shown as a montage in (e).
Supplementary Figure 6 Tracking accuracy.
(a) Performance metrics of the tracking routine from an example dataset plotting the position of the galvanometric mirrors together with the position of the fish, the distance of the head of the fish to the center of the FOV and the swimming speed. (b) Heat map of aggregate data (used in Fig. 2a) showing the distance of the head of the fish to the center of the FOV as a function of swimming speed to illustrate how the tracking algorithm minimizes the movements of the galvanometric mirrors. (c) Comparison of the performance of online tracking vs. offline tracking as measured by the deviation of the distance of the head of the fish to the center of the FOV. The offline tracking has access to the entire image series and can apply better denoising and segmentation (based on background subtraction and spatial filtering in the frequency domain).
Supplementary Figure 7 Image quality assessment and performance of registration routine.
The heat maps show several measures of the data shown in Figure 2 as a function of swimming speed: (a) the features detected by the initial quality check; frames with less than 5 features were discarded (b) the scaling factor of the similarity transformation obtained from the registration; this parameter was used to censor frames which deviated by more than 0.1 from unit scale (c) the registration error (ssd) from the final registration step of the remaining frames. (d) The histogram shows the imaging frames that survived the quality check and the subsequent selections based on registration quality.
Supplementary Figure 8 Flowchart and performance of autofocus routine using the ETL.
(a) The electrically focus-tunable lens (ETL) can be controlled from the acquisition GUI for calibration of the system and optimizing the focus for each experimental run, reporter line and developmental stage used. It can also be run via an autofocus routine to enable, e.g., refocusing for long-term recordings or screens. The routine operates by computing a focus measure of each acquired frame. If this measure is lower than a threshold, additional focus measures are obtained from images acquired by varying the focal length of the ETL in both directions. A 2nd-degree polynomial is fitted to the obtained focus measures vs. the focal lengths that were acquired. If the residuals of the fit are small, the ETL is set to the focal length that resulted in the largest focus measure. If the residuals are large, the maximum of the fitted curve is used to set the ETL to the corresponding focal length. (b) To systematically demonstrate the performance of the autofocus algorithm, we moved an arena containing a larva tg(HuC:H2B-GCaMP6s) out of focus and let the focus routine adjust the focal length of the ETL to recover focus. In the panels beneath the flowchart, three representative cases are shown. The left panel shows the case in which a drop of the focus measure occurred after a downward movement of the stage at 7 seconds (cyan, left y-axis). The control currents (-80 to -130 mA) that were sent to the ETL to change focal length and regain focus are plotted in orange (right y-axis). The bottom graph displays the focus measure against the ETL position for 10 acquisitions acquired at different focal lengths. The large black dot shows the estimation of the optimal focal length that was then used to recover focus. The middle graph shows a case in which the estimated optimal focal length (large black dot) falls outside the range of measurement that were taken; still, focus was recovered by setting the ETL to the predicted focal length. The right panel shows an instance in which the arena movement was continuing. This resulted in the focal measures from the acquisitions shown in red together with the fitted function and the estimated focal length. Since the first term of the fitted polynomial (shown in red) was negative, the routine selected the maximum focal measure it found among its acquired images (as opposed to the estimate) to set the focal length of the ETL. (c) Fine-focusing of the ETL (mΑ control currents correspond to submicron steps in the focal length) on a larval brain. (d) To showcase the focal length range of the ETL, fish swimming in different depths in a 10 mm deep arena were brought in focus. The bottom frame focuses on the right fish located close to the surface (fish is tilted). The top frame was acquired 200 ms later and focused on the left fish swimming at a greater depth.
Supplementary Figure 9 Additional analysis of neuronal activity during spontaneous swimming.
(a) Correlation plot of the fluorescent signal changes against the swimming speed in the anterior hindbrain (ant. hind), posterior hindbrain (post. hind), the medial midbrain (med. mid), and the optic tectum (OT) from the ROIs shown in Figure 1c. As compared to the other brain regions, OT showed lower correlations with speed. The plots in the right column show the same analysis for an animal with panneuronal expression of GFP instead of GCaMP6s (tg(HuC:Gal4;UAS:GFP x Ath5:Gal4) in which no correlated regions (more than 4 connected voxels) were found at the same threshold (p<10-5). b) Cluster analysis complementing the regression analysis shown in Figure 1c also identified highly correlated signal changes in the the anterior and posterior hindbrain regions (e.g., yellow cluster when searching for maximally 8 clusters). (c) Correlation plots vs. swimming speed of the fluorescent signal detected in the left and right cerebellum (ce) (and both sides combined) from a fish with selective expression of GCaMP6s in only the granule cells of the cerebellum (tg(gSA2AzGFF152B;UAS:GCaMP6s) corresponding to the experiment shown in Figure 1d.
Supplementary Figure 10 Different designs of the NeuBtracker arena.
(a) The default arena design consists of two CNS-milled plastic (or metal) parts, holding between them a glass coverslip of 9 mm in diameter (1 mm height) that can be easily exchanged between experiments. Shown here is a version used in the experiments of Figures 1-3 that has a central divider separating the arena in two compartments into which compounds can be delivered via diffusion from two quadratic reservoirs (b) The picture shows the illuminated arena. (c-e) The system allows for long-term recording over several hours in which case the water level (75-100 μL) can be kept constant by compensating for evaporation. This can be easily achieved via a syringe pump (green) as shown in the photograph. (f) A 150 mm travel path x-stage is incorporated into the system to move a (g) commercial multiwell-slides with 18 holes (5 mm diameter each) positioned by a custom-made 3D printed holder. We showcase the use of this configuration for sorting zebrafish based on their expression level of GCaMP5g (+ and – in the upper fluorescent image).
Supplementary Figure 11 Further characterization of neurobehavioral effects of cadaverine.
(a) Fluorescent signal changes recorded in the olfactory epithelium in the experiment shown in Figure 1e,f plotted here continuously over the entire duration of the experiment together with the distance of the fish to the cadaverine port at each point in time (color coding as in Figure 1f). (b) Control experiment in which water was injected instead of cadaverine with identical duration and settings to the experiment shown in Figure 1f,g. After the end of the 360 second baseline observation period, cadaverine was injected. Data are plotted analogously to Figure 1 and a. White scalebar represents 200 μm. (c) Additional cadaverine stimulation experiment run with a longer baseline than that shown in Figure 1f and showing again multiple visits to the cadaverine port with subsequent activations of oe. (d) The behavioral analyses show the median and the interquartile range of the mean duration of visits, as well as the mean swimming velocity in the left compartment before and after Cadaverine (Cad) delivery (100 μM). Cad reached the compartment via diffusion from a reservoir on the rim of the arena wall connected to the arena via a thin conduit (Wilcoxon matched-pairs signed rank tests, p = 0.0781, two-tailed (mean duration) and p = 0.023, two-tailed. (e) Representative heat maps of the behavioral traces for 3 different individuals during 2 minutes before (left) and after (right) delivery of cadaverine to the left compartment (purple shading in d). Black scalebar represents 2 mm.
Supplementary Figure 12 Comparison of light-dependent pineal complex activity in freely moving and restrained animals.
(a) Fluorescence signal changes obtained on NeuBtracker in MicroFixed configuration corresponding to the behavioral data shown in Figure 3 b,b’. The signal from the different ROIs is averaged over 4 animals with bounds of one standard deviation in dashed lines (fitted time constant: 11.9 ± 1.1 seconds.) (b) Single plane through pc (two-photon microscopy; inset shows magnification) as anatomical reference (c) Staining with the neural activity marker phosphorylated ERK (p-ERK, red), total ERK (t-ERK, green) and their ratio (p-ERK/t-ERK, cyan) from a fish imaged on NeuBtracker with ten cycles of 50 s ON/10 s OFF for 10 min. (d) Fluorescent signal changes (green traces) in the pineal complex (pc) during two cycles of light ON/OFF stimulation selected from the 10 cycles shown as average in Figure 3a from a short period in which the larva tg(gSA2AzGFF49A;UAS:GCaMP7a, 6 dpf) was relatively stationary (right) compared with a phase with swimming activity (left side) as seen from the plot of the concurrently recorded x- and y-position of the larva and its swimming speed (middle panel). The bottom panel shows as a reference the pc signal obtained from a restrained larva (embedded in agar, black trace). (e,f) To confirm the quality of image registration and control for any instrument-dependent effects on the signal changes, we performed the same experiment but using larvae expressing GFP tg(HuC:Gal4;UAS:eGFP) instead of a calcium indicator either freely swimming (e) or embedded in agar (f). The fluorescent signal traces are plotted from the ROIs indicated in the inset in (f). Scalebars represent 200 μm.
Supplementary Figure 13 Alternative optical paths for using NeuBtracker for imaging and stimulation.
(a) Simplified schematic showing additional options for optical ports in NeuBtracker here shown in MicroFixed configuration. (b) Example of coupling a 488 nm laser instead of the LED as an excitation light source. Efficient excitation of a GCAMP6 tg(HuC:H2B-GCaMP6s) could also be achieved with a laser but speckle patterns were apparent (which could be removed by a rotating diffuser or alternatively used for structured illumination approaches). (c) The transparent bottom of the arena and the use of an NIR illumination ring rather than a table afford direct coupling of a video projector to provide visual stimulation to the animal. (d) A 405 nm laser was coupled via the galvanometric mirrors to provide a visual stimulus to the animal. During periods in which the laser spot was moved to repeatedly approach the head, the fish swam a longer distance than during periods in which the 405 nm laser was off. This was the case with and without a 488 nm laser turned on in the background. These data are consistent with data we obtained from fish immobilized in agar in which illumination of the head with the 405 nm laser lead to an increase in the number of specific tail deflections that can be characterized as C-turns (indicative of aversion) and J-turns by videography conducted with an IR camera. Individual counts from 6 individuals are shown with median and interquartile range. (e) In this experiment, the 405 nm laser spot was moved in a semi-circular path in a left-right-left trajectory in front of the head of a tg(gSA2AzGFF49A;UAS:GCaMP7a, 7 dpf) larva as indicated by color-coding for relative position. Neuronal activations were elicited in the optic tectum contralateral to the relative location of the laser spot (the color overlay over the co-registered frames in gray shows the ratio of the average fluorescence between frames in which the laser spot was located on the left and frames in which it was positioned on the right relative to the location of the fish head). The plot (median and interquartile range) shows a significant difference (p < 0.01, Wilcoxon matched-pairs signed rank test) of the ratio of the average fluorescence intensity of the left and right OT in frames with laser spots presented on the left (green) or right (red) of the fish respectively.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 and Supplementary Table 1.
Life Sciences Reporting Summary
Life Sciences Reporting Summary.
Supplementary Software
NeuBtracker software suite - data acquisition and processing routines
NeuBtracker: an open-source imaging platform for interrogating neurobehavioral dynamics in freely behaving fish
Overview of the design, features, data acquisition and processing pipeline of NeuBtracker
Rights and permissions
About this article
Cite this article
Symvoulidis, P., Lauri, A., Stefanoiu, A. et al. NeuBtracker—imaging neurobehavioral dynamics in freely behaving fish. Nat Methods 14, 1079–1082 (2017). https://doi.org/10.1038/nmeth.4459
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4459
This article is cited by
-
Large-scale neural recordings call for new insights to link brain and behavior
Nature Neuroscience (2022)
-
Fast, efficient, and accurate neuro-imaging denoising via supervised deep learning
Nature Communications (2022)
-
Imaging whole nervous systems: insights into behavior from worms to fish
Nature Methods (2019)
-
Attraction of posture and motion-trajectory elements of conspecific biological motion in medaka fish
Scientific Reports (2018)
-
A user-friendly herbicide derived from photo-responsive supramolecular vesicles
Nature Communications (2018)