Abstract
The stable expansion of tissue-specific stem cells in vitro has contributed to research on several organs. Alveolar epithelial type II (AT2) cells function as tissue stem cells in the lung, but robust models for studying human AT2 cells are lacking. Here we report a method for the efficient generation and long-term expansion of alveolar organoids (AOs) harboring SFTPC+ alveolar stem cells derived from human induced pluripotent stem cells (hiPSCs). hiPSC-derived SFTPC+ cells self-renewed, with transcriptomes and morphology consistent with those of AT2 cells, and were able to differentiate into alveolar epithelial type I (AT1)-like cells. Single-cell RNA-seq of SFTPC+ cells and their progenitors demonstrated that their differentiation process and cellular heterogeneity resembled those of developing AT2 cells in vivo. AOs were applicable to drug toxicology studies recapitulating AT2-cell-specific phenotypes. Our methods can help scientists overcome the limitations of current approaches to the modeling of human alveoli and should be useful for disease modeling and regenerative medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
14 November 2017
In the version of this article initially published, there were errors in the Online Methods section. Specifically, incorrect concentrations were given for 8-Br-cAMP, 3-isobutyl-1-methylxanthine, and KGF, as used for induction and passage of alveolar stem cells in fibroblast-dependent organoids. The errors have been corrected in the HTML and PDF versions of the article.
References
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Li, Z. et al. 3D culture supports long-term expansion of mouse and human nephrogenic progenitors. Cell Stem Cell 19, 516–529 (2016).
Mason, R.J. & Williams, M.C. Type II alveolar cell. Defender of the alveolus. Am. Rev. Respir. Dis. 115, 81–91 (1977).
Barkauskas, C.E. et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013).
Tsuji, T., Aoshiba, K. & Nagai, A. Alveolar cell senescence in patients with pulmonary emphysema. Am. J. Respir. Crit. Care Med. 174, 886–893 (2006).
Bueno, M. et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J. Clin. Invest. 125, 521–538 (2015).
Whitsett, J.A., Wert, S.E. & Weaver, T.E. Diseases of pulmonary surfactant homeostasis. Annu. Rev. Pathol. 10, 371–393 (2015).
Desai, T.J., Brownfield, D.G. & Krasnow, M.A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014).
Dobbs, L.G., Williams, M.C. & Gonzalez, R. Monoclonal antibodies specific to apical surfaces of rat alveolar type I cells bind to surfaces of cultured, but not freshly isolated, type II cells. Biochim. Biophys. Acta 970, 146–156 (1988).
Borok, Z. et al. Modulation of T1α expression with alveolar epithelial cell phenotype in vitro. Am. J. Physiol. 275, L155–L164 (1998).
Green, M.D. et al. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 29, 267–272 (2011).
Longmire, T.A. et al. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398–411 (2012).
Schmeckebier, S. et al. Keratinocyte growth factor and dexamethasone plus elevated cAMP levels synergistically support pluripotent stem cell differentiation into alveolar epithelial type II cells. Tissue Eng. Part A 19, 938–951 (2013).
Ghaedi, M. et al. Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J. Clin. Invest. 123, 4950–4962 (2013).
Huang, S.X. et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32, 84–91 (2014).
Gotoh, S. et al. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Rep. 3, 394–403 (2014).
McCauley, K.B. et al. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell 20, 844–857 (2017).
Rawlins, E.L., Clark, C.P., Xue, Y. & Hogan, B.L. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–3745 (2009).
Konishi, S. et al. Directed induction of functional multi-ciliated cells in proximal airway epithelial spheroids from human pluripotent stem cells. Stem Cell Rep. 6, 18–25 (2016).
Morrisey, E.E. & Hogan, B.L. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell 18, 8–23 (2010).
Tsao, P.N. et al. γ-Secretase activation of Notch signaling regulates the balance of proximal and distal fates in progenitor cells of the developing lung. J. Biol. Chem. 283, 29532–29544 (2008).
Karrer, H.E. The ultrastructure of mouse lung; general architecture of capillary and alveolar walls. J. Biophys. Biochem. Cytol. 2, 241–252 (1956).
Miklavc, P. et al. Actin coating and compression of fused secretory vesicles are essential for surfactant secretion—a role for Rho, formins and myosin II. J. Cell Sci. 125, 2765–2774 (2012).
Frank, D.B. et al. Emergence of a wave of Wnt signaling that regulates lung alveologenesis by controlling epithelial self-renewal and differentiation. Cell Rep. 17, 2312–2325 (2016).
Liu, Y., Martinez, L., Ebine, K. & Abe, M.K. Role for mitogen-activated protein kinase p38α in lung epithelial branching morphogenesis. Dev. Biol. 314, 224–235 (2008).
Chung, C. et al. Hippo-Foxa2 signaling pathway plays a role in peripheral lung maturation and surfactant homeostasis. Proc. Natl. Acad. Sci. USA 110, 7732–7737 (2013).
Lin, S., Perl, A.K. & Shannon, J.M. Erm/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. J. Biol. Chem. 281, 16716–16726 (2006).
Rosenberg, E. et al. Members of the C/EBP transcription factor family stimulate expression of the human and rat surfactant protein A (SP-A) genes. Biochim. Biophys. Acta 1575, 82–90 (2002).
Gonzales, L.W., Guttentag, S.H., Wade, K.C., Postle, A.D. & Ballard, P.L. Differentiation of human pulmonary type II cells in vitro by glucocorticoid plus cAMP. Am. J. Physiol. Lung Cell. Mol. Physiol. 283, L940–L951 (2002).
Watanabe, K. et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 (2007).
Shu, W. et al. Wnt/β-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev. Biol. 283, 226–239 (2005).
Jain, R. et al. Plasticity of Hopx+ type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 6, 6727 (2015).
Xu, Y. et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1, e90558 (2016).
Cunningham, A.C. et al. Constitutive expression of MHC and adhesion molecules by alveolar epithelial cells (type II pneumocytes) isolated from human lung and comparison with immunocytochemical findings. J. Cell Sci. 107, 443–449 (1994).
Van der Velden, J.L., Bertoncello, I. & McQualter, J.L. LysoTracker is a marker of differentiated alveolar type II cells. Respir. Res. 14, 123 (2013).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Ridsdale, R. & Post, M. Surfactant lipid synthesis and lamellar body formation in glycogen-laden type II cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 287, L743–L751 (2004).
Suzuki, A. et al. Single-cell analysis of lung adenocarcinoma cell lines reveals diverse expression patterns of individual cells invoked by a molecular target drug treatment. Genome Biol. 16, 66 (2015).
Du, Y., Guo, M., Whitsett, J.A. & Xu, Y. 'LungGENS': a web-based tool for mapping single-cell gene expression in the developing lung. Thorax 70, 1092–1094 (2015).
Bedrossian, C.W., Warren, C.J., Ohar, J. & Bhan, R. Amiodarone pulmonary toxicity: cytopathology, ultrastructure, and immunocytochemistry. Ann. Diagn. Pathol. 1, 47–56 (1997).
Funayama, M. et al. A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2–q13.1. Ann. Neurol. 51, 296–301 (2002).
Herzig, M.C. et al. LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum. Mol. Genet. 20, 4209–4223 (2011).
Miklavc, P. et al. Surfactant secretion in LRRK2 knock-out rats: changes in lamellar body morphology and rate of exocytosis. PLoS One 9, e84926 (2014).
Fuji, R.N. et al. Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci. Transl. Med. 7, 273ra15 (2015).
Sirianni, F.E., Chu, F.S. & Walker, D.C. Human alveolar wall fibroblasts directly link epithelial type 2 cells to capillary endothelium. Am. J. Respir. Crit. Care Med. 168, 1532–1537 (2003).
Lee, J.H. et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156, 440–455 (2014).
Whitsett, J.A. & Alenghat, T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16, 27–35 (2015).
Fehrenbach, H. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir. Res. 2, 33–46 (2001).
Yamamoto, Y. & Gotoh, S. Methods of generating human pluripotent stem cell–derived alveolar stem cells and their expansion. Protoc. Exch. http://dx.doi.org/10.1038/protex.2017.097 (2017).
Thomson, J.A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Okita, K. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 (2011).
Okita, K. et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31, 458–466 (2013).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Fujino, N. et al. A novel method for isolating individual cellular components from the adult human distal lung. Am. J. Respir. Cell Mol. Biol. 46, 422–430 (2012).
Yamano, G. et al. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett. 508, 221–225 (2001).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank K. Osafune, K. Okita, K. Takahashi and S. Yamanaka (Center for iPS Cell Research and Application, Kyoto University, Kyoto, Japan) for providing iPSC lines. We thank F. Chen, H. Date and all the members of the Department of Thoracic Surgery, Kyoto University (Kyoto, Japan), for providing surgical specimens of human adult lung. We thank K. Okamoto-Furuta and H. Kohda (Center for Anatomical Studies, Kyoto University, Kyoto, Japan) for electron microscopy studies. We thank T. Horiuchi and K. Imamura (Department of Medical Genome Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Japan) for scRNA-seq. We thank K. Ando and K. Seyama (Juntendo University Faculty of Medicine and Graduate School of Medicine, Tokyo, Japan) for technical guidance on the isolation of human adult alveolar epithelial cells. We thank N. Inagaki and D. Tanaka (Department of Diabetes, Endocrinology and Nutrition, Kyoto University, Kyoto, Japan) for kindly providing anti-human ABCA3 antibody, and M. Hagiwara, Y. Okuno and all the members of Medical Research Support Center, Kyoto University (Kyoto, Japan), for helpful discussion and supporting use of Gene Spring software and fluorescence studies. We also thank Y. Maeda and A. Inazumi for technical support. This work was supported by MEXT of Japan (Grant-in-Aid for Scientific Research (KAKENHI) numbers 15K21114 and 17H05084 to S.G., 15H04318 and 16H06279 to Y.S., and 22249031 and 15H02537 to M.M.), the Japan Agency for Medical Research and Development (grants KU-A032, 16bm0704008h0001 and 17bm0704008h0002 to S.G.), KANAE Foundation for the Promotion of Medical Science (to S.G.), Takeda Science Foundation (to S.G.), and in part by Daiichi Sankyo (research grant to T.H. and M.M.).
Author information
Authors and Affiliations
Contributions
Y.Y. and S.G. conceived and designed the study. Y.Y., S.G., Y.K., M.S., S.K., S.I., N.S. and T.N. conducted the experiments. Y.Y., S.G., M.S., T.K. and Y.S. analyzed the data. Y.Y. and S.G. wrote the manuscript after fruitful discussion with and under the supervision of M.S., H.M., S.M., I.I., T.H., T.K., Y.S. and M.M.
Corresponding author
Ethics declarations
Competing interests
Kyoto University has applied for a patent related to the method of alveolar cell differentiation from induced pluripotent stem cells discussed in this paper.
Additional information
Division of Genome Biology, National Cancer Center Research Institute, Tokyo, Japan
Integrated supplementary information
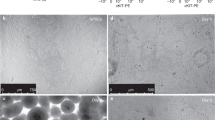
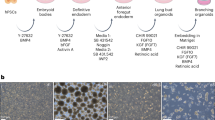
Supplementary Figure 1 Efficient generation of AOs via isolated NKX2-1+ lung progenitor cells.
(a) Optimization of the medium for expansion of NKX2-1+ ventralized anterior foregut endoderm (VAFE) cells on day 21. IBMX, 3-isobutyl-1-methylxanthine. (b) Immunofluorescence imaging of NKX2-1+ cell expansion under each of the medium conditions described in (a) on day 21 (n = 3 repeated experiments, each). Scale bar, 100 μm. (c) Induction efficiency of NKX2-1+ cells on day 21 under each of the medium conditions calculated by the averaged ratio of the NKX2-1+ cells to the total cells in 10 randomly selected fields. CFKD treatment was found to be significantly more effective than FGF10 treatment by Kruskal-Wallis test combined with Dunn’s post hoc multiple comparison test. *P < 0.05. Error bars indicate the mean ± s.e.m (n = 3 cell cultures) B2-3 hiPSC line was used. (d) Optimizing the duration of expansion of NKX2-1+ VAFE cells in CFKD medium. (e) RT-qPCR of SFTPC for each duration of NKX2-1+ VAFE cells expansion after day 14. The expression of SFTPC in AOs after one-week CFKD treatment was significantly greater than that in untreated cells by Kruskal-Wallis test combined with Dunn’s post hoc multiple comparison test. *P < 0.05. The expression of SFTPC was normalized to that of ACTB. The expression of SFTPC in human fetal lung was set at 1. Error bars indicate the mean ± s.e.m (n = 3 cell cultures, each). B2-3 hiPSC line was used. (f) A flow cytometric analysis (n = 5 repeated experiments) of the CPM-based sorting of cells into three cell populations (CPMhi, CPMlo and CPM-). Mouse IgG1 isotype was used as a negative control. H9 ESC line was used. (g) Immunofluorescence images of NKX2-1+ cells of cytospin samples in sorted CPMhi, CPMlo and CPM- cells (n = 3 repeated experiments). H9 ESC line was used. Scale bar, 100 μm. (h) The ratio of NKX2-1+ cells in cells sorted based on CPM in multiple PSC lines. Error bars indicate the mean ± s.e.m (n = 3 cell cultures). The ratio of NKX2.1+ cells in CPMhi cells was found to be significantly greater than that in CPM- cells by Friedman test combined with Dunn’s post hoc multiple comparison test. *P < 0.05. (i) A histogram of the accumulation of LysoTracker Red in SFTPC+ cells in AOs. Numbers indicate the mean ± s.e.m. (n = 3 independent organoids). (j) A gating strategy for flow cytometric analysis of AOs. (k, l) Flow cytometry analysis (k) and graphs (l) of induction efficiency of SFTPC+ cells forming AOs cocultured with various fibroblasts. Numbers and error bars indicate the mean ± s.e.m (n = 5 independent organoids, each). (m) An immunofluorescent image of AOs stained with anti-GFP (SFTPC+ cells), anti-EpCAM (epithelial cells), and anti-CD31 (endothelial cells) (n = 3 repeated experiments). Scale bar, 25 μm.
Supplementary Figure 2 RT-qPCR of various airway epithelial cell markers on day 35 in multiple PSC lines.
The following markers were used: FOXJ1 and SNTN for ciliated cells, MUC5AC and SPDEF for goblet cells, TP63 and KRT5 for basal cells, and SCGB1A1 and SCGB3A2 for club cells. The expression of each gene was normalized to that of ACTB. The expression in human fetal trachea was set at 1. Human adult trachea and liver were used as additional controls. Error bars indicate the mean ± s.e.m (n = 3 independent organoids). *P < 0.05 in the comparison of the gene expression of CPMhi cell-derived organoids and CPMhi progenitor cells; # P < 0.05 and ## P < 0.01 in the comparison of the gene expression of organoids derived from CPMhi and CPM- cells. Two-tailed unpaired t- test (df = 4) was performed independently.
Supplementary Figure 3 AT2-cell-associated gene expression profile of hiPSC-derived SFTPC+ cells.
(a) The numbers of significantly upregulated genes in hiPSC-derived SFTPC+ cells relative to CPMhi progenitor and SFTPC- cells in microarray data (n = 3 independent experiments for each condition, >2-fold, P < 0.05, ANOVA with Tukey’s post hoc) (GSE90590). (b) A pie chart showing GO term distribution for the 740 upregulated genes in hiPSC-derived SFTPC+ cells. (c) A list of 75 GO terms (“cellular component”, [33 terms]; “biological process”, [38 terms]; and “molecular function”, [4 terms]) found to be significantly enriched in SFTPC+ cell transcriptomes by Fisher’s exact test (P < 0.05) (GSE90590). The red bars correspond to GO terms in (b). (d) Heat maps of the gene expression annotated with the GO terms of AT2 cell function in comparison of hiPSC-derived CPMhi progenitor cells and SFTPC+ and SFTPC- cells. (e) A list of the top 25 upregulated genes in hiPSC-derived SFTPC+ cells. An immunofluorescent analysis was performed to detect expression of the genes shown in red. See also Supplementary Fig. 4 and Supplementary Table 1. B2-3 hiPSC line was used in all of the experiments.
Supplementary Figure 4 Representative immunofluorescence images from a microarray study showing upregulated AT2 marker expression in hiPSC-derived SFTPC+ cells.
SLC34A2, PGC, NAPSA, LPCAT1, LRRK2 and CTSH were expressed in hiPSC-derived SFTPC+ or DCLAMP+ cells and fetal and adult lung tissues (n = 3 collected images for each sample). Scale bars, 25 μm. B2-3 cell line was used.
Supplementary Figure 5 scRNA-seq analysis of differentiation from CPMhi progenitor cells to SFTPC+ cells.
(a) The data of the sequenced reads in CPMhi cells on day 14 and 21 and EpCAM+ cells on day 26. (b) The numbers of detected reads of RNA spike-in controls added to single-cell lysates of CPMhi cells on day 14 and 21, and EpCAM+ cells, respectively. The detected reads (log10) increased according to the number of estimated copies of spike-in controls mixed in each lysate (shown in the text box). Error bars indicate the mean ± 2 s.d. (c) The correlation between the mean expression of each of 24,104 genes in single cells and 200 bulk cells in CPMhi cells on day 14 and 21 and EpCAM+ cells, respectively. r indicates Pearson’s correlation coefficient. (d) The correlation among the mean expression of each of 24,104 genes in CPMhi cells on day 14 and 21 and EpCAM+ single cells. r indicates Pearson’s correlation coefficient. (e) PCA of the gene expression profiles of 206 single cells on 17,873 genes. Three-dimensional image is shown in Figure 2c (GSE90813).
Supplementary Figure 6 Fibroblast-free (FF) AO induction and scRNA-seq analysis of FF-SFTPC+ cells.
(a) Representative plots of the flow cytometric analysis of FF-AOs treated with CHIR99021 (3 μM), SB431542 (10 μM), their combination (2i), and vehicle control on day 28. Numbers indicate the mean ± s.e.m (n = 3 independent organoids, each). (b) The data of the sequenced reads in SFTPC+ single-cells induced under FF condition with 2i treatment (FF-SFTPC+ cells). (c) The numbers of detected reads of RNA spike-in controls of FF-SFTPC+ single cell lysates. The detected reads (log10) increased according to the number of estimated copies of spike-in controls in each lysate (shown in the text box). Error bars indicate the mean ± 2 s.d. (d) The correlation between the mean expression of each of 24,104 genes in single cells and 200 bulk cells in FF-SFTPC+ cells. r indicates Pearson’s correlation coefficient. (e) The correlation among the mean expression of each of 24,104 genes in FF-SFTPC+ cells, CPMhi progenitor cells on day 21, and SFTPC+ cells induced under fibroblast-dependent condition (FD-SFTPC+ cells) on day 26. r indicates Pearson’s correlation coefficient. (f) Hierarchical clustering analysis of single cell transcriptomes of 85 CPMhi progenitor cells (day 21), 70 FF-SFTPC+ cells, and 50 FD-SFTPC+ cells (GSE90813) based on the representative lung epithelial cell markers described in Supplementary Table 3. B2-3 hiPSC line was used for all of the experiments.
Supplementary Figure 7 Stable proliferation of hiPSC-derived FD-SFTPC+ cells.
(a) The self-renewal rate of FD-SFTPC+ cells in FD-AOs. A representative flow cytometric analysis is shown. P1, Passage number 1. “P1 FD-AO” indicates AOs derived from P0 FD-SFTPC+ cells. The values were calculated as the ratio of SFTPC+ cells to EpCAM+ cells. Error bars indicate the mean ± s.e.m (n = 3 independent organoids, each). (b) Linear increase in the population doubling level (PDL) of FD-SFTPC+ cells was observed. PDL was calculated as log10 (N/N0) x 3.33. N is the number of EpCAM+ cells. N0 is the number of SFTPC+ cells and was fixed at 1.0 x 104 in the present study. Linear regression analysis with Pearson’s correlation coefficient was used to examine relationship between the increase in the culture period and PDL. r, Pearson’s correlation coefficient. Error bars indicate the mean ± s.e.m (n = 3 independent organoids, each). (c) Normal karyotypes of P0 and P5 FD-SFTPC+ cells. (d) Confocal immunofluorescence imaging of P3 FD-AOs (n = 5 collected images). Scale bar, 25 μm. (e) Confocal immunofluorescence imaging of SFTPC-PDPN+AQP5+ AT1-like cells in P3 FD-AOs (n = 5 collected images). Scale bar, 25 μm. (f) Flow cytometric analysis of FD-AOs for the quantification of maturation of lamellar bodies with calculation of the mean signal intensity of LysoTracker. Error bars indicate the mean ± s.e.m (n = 3 independent organoids, each). *P < 0.05, Kruskal-Wallis test combined with Dunn’s post hoc multiple comparison test. B2-3 hiPSC line was used for all of the experiments.
Supplementary Figure 8 Global gene expression pattern of expanded hiPSC-derived FD-SFTPC+ cells.
(a) A non-biased PCA of the transcriptomes of P0, P2 and P5 hiPSC-derived FD-SFTPC+ cells, CPMhi progenitor cells, hiPSC-derived proximal airway epithelial cells (PAECs) (n = 3 independent experiments, each) and primary isolated AT2 cells (n = 3 donors) (GSE90591). See also Figure 4f. (b) Non-biased hierarchical clustering of the transcriptomes of P0, P2 and P5 hiPSC-derived FD-SFTPC+ cells, CPMhi progenitor cells, PAECs (n = 3 independent experiments, each) and primary isolated AT2 cells (n = 3 donors). (c) Hierarchical clustering of each transcriptome (columns) based on representative lung epithelial cell markers described in Supplementary Table 3 (rows). B2-3 hiPSC line was used in all experiments, except for parental 201B7 hiPSC-derived PAECs.
Supplementary Figure 9 A comparison of the transcriptomes of expanded hiPSC-derived FD-SFTPC+ cells with those of human adult AT2 cells.
(a) A Venn diagram showing 6142 adult AT2-specific genes listed by analyzing the transcriptomes of three conditions: primary adult AT2 cells isolated from human lung (n = 3 donors); P0, P2 and P5 hiPSC-derived FD-SFTPC+ cells (n = 3 independent experiments, each); CPMhi progenitor cells (n=3 biological replicates) (>2-fold upregulated genes, P < 0.05, ANOVA corrected by Benjamini-Hochberg FDR with Tukey’s post hoc). See also Supplementary Table 4. (b) A list of top 20 GO terms that were significantly annotated with adult AT2 specific genes (P < 0.05, Fisher’s exact test). See also Supplementary Table 5.
Supplementary Figure 10 Expansion of alveolar stem cell populations by using LysoTracker without genetic reporter cell lines.
(a) A schematic illustration of expansion of alveolar stem cells by using LysoTracker. (b) Representative histograms of LysoTracker Red for each cell component of FD-AOs (EpCAM+SFTPC+ cells, EpCAM+SFTPC- cells, and mesenchymal cells). (c) RT-qPCR analysis of alveolar epithelial cell markers in FD-AOs derived from EpCAM+LysoTrackerhi cell population in five PSC lines compared with those in P1 FD-AOs derived from FD-SFTPC+ cells using genetic reporter (B2-3) -based cell sorting. The expression human fetal lung was set at 1. Error bars indicates the mean ± s.e.m (n=3 independent organoids, each). (d) A TEM image of lamellar bodies in P2 FD-AO derived from EPCAM+LysoTrackerhigh cells (n = 3 collected images). Scale bar, 2 μm. (e) Immunofluorescence images of AT2 markers in P2 FD-AO derived from EPCAM+LysoTrackerhi cells (n = 5 collected images). B2-3 and 648A1 hiPSC lines were used in (b) and (d) (e), respectively.
Supplementary Figure 11 Cellular heterogeneity of hiPSC-derived FD-SFTPC+ cells.
(a) A flow cytometric analysis of PDPN and SFTPC expression in EPCAM+ gated cells of P0, P1, P2, P5 and P6 FD-AOs, respectively. The values indicate the percentage of each population, mean ± s.e.m (n = 3 independent organoids, each). (b) The ratio of PDPN+/- cells in FD-SFTPC+ cells in each passage. *P < 0.05, Kruskal-Wallis test combined with Dunn’s post hoc multiple comparison test. Error bars indicate the mean ± s.e.m (n = 3 independent organoids, each). (c) TEM images of isolated P2 FD-SFTPC+ cells. The cells were morphologically classified into the three following three types: “immature type”, cells with large glycogen vacuole-like structures without lamellar bodies; “early type”, cells with the glycogen vacuoles with small lamellar bodies; and “late type”, cell with small or no glycogen vacuoles with well-structured lamellar bodies (n = 3 repeated experiments). B2-3 hiPSC line was used for all of the experiments.
Supplementary Figure 12 Validation of the scRNA-seq of hiPSC-derived FD-SFTPC+ cells.
(a) The data of the sequenced reads in P0, P2 and P5 FD-SFTPC+ single-cells. (b) The numbers of detected reads of RNA spike-in controls added to P0, P2 and P5 FD-SFTPC+ single-cell lysates, respectively. The detected reads (log10) increased according to the estimated copies of each spike-in control mixed into each lysate (shown in the text box). Error bars indicate the mean ± 2 s.d. (c) Correlation between the mean expression of each of 24,104 genes in single cells and 200 bulk cells in P0, P2 and P5 FD-SFTPC+ cells, respectively. r indicates Pearson’s correlation coefficient. (d) Correlation among the mean expression of each of 24,104 genes in P0, P2 and P5 FD-SFTPC+ single cells. r indicates Pearson’s correlation coefficient.
Supplementary Figure 13 Heterogeneity of hiPSC-derived FD-SFTPC+ cells in single-cell analysis.
(a) Immunofluorescence images of several AT1 markers (PDPN, CAV1, AGER and HOPX) costained with GFP (SFTPC+ cells) in P3-AOs (n = 3 collected images). Error bars, 25 μm. (b) RT-qPCR of AT1 and AT2 marker expression in two distinct representative cell populations at single-cell resolution. “Positive” and “negative” indicate cell populations that were positive or negative for all AT1 markers of HOPX, PDPN, CAV1 and AGER, respectively. Error bars indicate the mean ± s.e.m. n = 13 and 15 single cells were analyzed as “positive” and “negative” population, respectively. *P < 0.05, **P < 0.01, N.S., not significant by two-tailed unpaired t-test (df = 26). (c) The component ratio of AT1 marker-based classification of P0, P2 and P5 FD-SFTPC+ cells. “High”, “moderate” and “low” indicate the cells expressing three/four, two and one/none of four AT1 markers (HOPX, PDPN, CAV1 and AGER), respectively. N.S., not significant by chi-squared test. (d) A 3D view of the PCA of 138 FD-SFTPC+ single cells based on 16,906 genes (GSE90813). (e) A 2D scattergram of the PCA depicted in (c).
Supplementary Figure 14 The cellular heterogeneity of developing AT2 cells, depicted by novel gene sets identified on the basis of AT1-marker-based clustering of hiPSC-derived FD-SFTPC+ cells.
(a) GO terms in each gene cluster shown in Figure 5e. Fisher’s exact test was used for the analysis (P < 0.05). (b) Validation using murine a single-cell transcriptome database (GSE52583) of Sftpc+ cells in E18.5 and P107 murine lung37. A hierarchical clustering of 106 single cells was performed using 134 of 144 genes that corresponded to the human genes described in Figure 5e. The color bars at the left and right of the heat map indicate the developmental stages of murine Sftpc+ cells and AT1-marker based classification of Sftpc+ cells defined in Figure 5b, respectively. FRKM, fragments per kilobase of exon per million mapped reads.
Supplementary Figure 15 Transcriptomic analysis of FD-SFTPC+ cells treated with GNE7915 and amiodarone.
A list of the top 20 GO terms annotated with three distinct gene sets comparing the transcriptomes of GNE7915- or amiodarone-treated P3 FD-SFTPC+ cells with that of vehicle control, respectively (GSE99939) (n = 3 independent experiments, each). A Venn diagram shows the number of genes in each set. See also Supplementary Tables 7, 8. B2-3 cell line was used in all of the experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Tables 3, 5, 10 and 11.
Life Sciences Reporting Summary
Life Sciences Reporting Summary
Supplementary Protocol
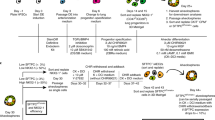
Methods of generating human pluripotent stem cell (PSC)-derived alveolar stem cells and their expansion
Supplementary Table 1
Genes that were significantly upregulated inhiPSC-derived SFTPC+ cells compared with expression in SFTPC– cells and CPMhi progenitor cells, as determined by microarray
Supplementary Table 2
Genes associated with preconditioning of VAFE cells
Supplementary Table 4
Genes that were significantly upregulated in adult AT2 cells compared with their expression in FD-SFTPC+ cells
Supplementary Table 6
Genes with significantly different expression between AT1-marker-high and AT1-marker-low cells, as determined by single-cell RNA-seq
Supplementary Table 7
Genes whose expression changed in GNE7915-treated or amiodarone-treated P3 FD-SFTPC+ cells
Supplementary Table 8
GO terms associated with genes that responded to treatment with GNE7915 and/or amiodarone
Supplementary Table 9
Performance of the methods in the present study compared with that in the previous study
Rights and permissions
About this article
Cite this article
Yamamoto, Y., Gotoh, S., Korogi, Y. et al. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat Methods 14, 1097–1106 (2017). https://doi.org/10.1038/nmeth.4448
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4448
This article is cited by
-
Organoids in ovarian cancer: a platform for disease modeling, precision medicine, and drug assessment
Journal of Cancer Research and Clinical Oncology (2024)
-
Lung Organoids: Systematic Review of Recent Advancements and its Future Perspectives
Tissue Engineering and Regenerative Medicine (2024)
-
Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant
Nature Communications (2024)
-
Human alveolar hydrogels promote morphological and transcriptional differentiation in iPSC-derived alveolar type 2 epithelial cells
Scientific Reports (2023)
-
Directed differentiation of mouse pluripotent stem cells into functional lung-specific mesenchyme
Nature Communications (2023)