Abstract
We engineered covalently circularized nanodiscs (cNDs) which, compared with standard nanodiscs, exhibit enhanced stability, defined diameter sizes and tunable shapes. Reconstitution into cNDs enhanced the quality of nuclear magnetic resonance spectra for both VDAC-1, a β-barrel membrane protein, and the G-protein-coupled receptor NTR1, an α-helical membrane protein. In addition, we used cNDs to visualize how simple, nonenveloped viruses translocate their genomes across membranes to initiate infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bayburt, T.H., Carlson, J.W. & Sligar, S.G. J. Struct. Biol. 123, 37–44 (1998).
Bayburt, T.H., Grinkova, Y.V. & Sligar, S.G. Nano Lett. 2, 853–856 (2002).
Ritchie, T.K. et al. Methods Enzymol. 464, 211–231 (2009).
Hagn, F., Etzkorn, M., Raschle, T. & Wagner, G. J. Am. Chem. Soc. 135, 1919–1925 (2013).
Raschle, T. et al. J. Am. Chem. Soc. 131, 17777–17779 (2009).
Grinkova, Y.V., Denisov, I.G. & Sligar, S.G. Protein Eng. Des. Sel. 23, 843–848 (2010).
Raschle, T., Hiller, S., Etzkorn, M. & Wagner, G. Curr. Opin. Struct. Biol. 20, 471–479 (2010).
Antos, J.M. et al. J. Biol. Chem. 284, 16028–16036 (2009).
Chen, I., Dorr, B.M. & Liu, D.R. Proc. Natl. Acad. Sci. USA 108, 11399–11404 (2011).
Hogle, J.M. Annu. Rev. Microbiol. 56, 677–702 (2002).
Tuthill, T.J., Groppelli, E., Hogle, J.M. & Rowlands, D.J. Viruses 343, 43–89 (2010).
Marsh, M. & Helenius, A. Cell 124, 729–740 (2006).
Racaniello, V.R. Fields Virology 6th edn. (eds. Knipe, D.M. & Howley, P.M.) 453–489 (Lippincott, Williams & Wilkens, 2013).
Mendelsohn, C.L., Wimmer, E. & Racaniello, V.R. Cell 56, 855–865 (1989).
Brandenburg, B. et al. PLoS Biol. 5, e183 (2007).
Tuthill, T.J., Bubeck, D., Rowlands, D.J. & Hogle, J.M. J. Virol. 80, 172–180 (2006).
Bubeck, D., Filman, D.J. & Hogle, J.M. Nat. Struct. Mol. Biol. 12, 615–618 (2005).
Strauss, M., Levy, H.C., Bostina, M., Filman, D.J. & Hogle, J.M. J. Virol. 87, 3903–3914 (2013).
Strauss, M. et al. J. Virol. 89, 4143–4157 (2015).
Maresso, A.W. et al. J. Biol. Chem. 282, 23129–23139 (2007).
Egloff, P. et al. Proc. Natl. Acad. Sci. USA 111, E655–E662 (2014).
Egloff, P., Deluigi, M., Heine, P., Balada, S. & Plückthun, A. Protein Expr. Purif. 108, 106–114 (2015).
Rasmussen, S.G. et al. Nature 477, 549–555 (2011).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Biol. Proced. Online 6, 23–34 (2004).
Tang, G. et al. J. Struct. Biol. 157, 38–46 (2007).
Hyberts, S.G., Milbradt, A.G., Wagner, A.B., Arthanari, H. & Wagner, G. J. Biomol. NMR 52, 315–327 (2012).
Delaglio, F. et al. J. Biomol. NMR 6, 277–293 (1995).
Mastronarde, D.N. J. Struct. Biol. 152, 36–51 (2005).
Li, X., Grigorieff, N. & Cheng, Y. J. Struct. Biol. 172, 407–412 (2010).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. Nat. Methods 9, 671–675 (2012).
Acknowledgements
The authors thank T. Raschle for his early contribution to this work and H. Chou, H. Arthanari and J. Ziarek for helpful discussions. The authors gratefully acknowledge the use of the services of the Taplin Mass Spectrometry Facility and the Center of Macromolecular Interactions (CMI). Addgene plasmid no. 75144 was a gift from D. Liu (Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts, USA). This work was supported by NIH grant 32GM113406 to M.L.N.; grant AI20566 to J.M.H.; and grants GM047467, GM075879 and AI037581 to G.W. G.W., A.P. and F.H. are supported by the Human Frontiers Science program. D.B. acknowledges support from the HHMI as a Gilliam Fellow. F.H. acknowledges support from the TUM Institute for Advanced Study cofunded by the European Commission as well as from the CIPSM excellence cluster and the DFG (SFB1035).
Author information
Authors and Affiliations
Contributions
The project was conceived by M.L.N. and G.W. Experiments were designed by M.L.N., D.B., G.W., M.S. and J.M.H. Experiments were carried out by M.L.N., D.B., M.S., Z.J.S., S.G. and S.H. The manuscript was written by M.L.N. and G.W. T.W., J.M.H., D.B., F.H., A.P. and S.G. contributed to discussions and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.N. and G.W. have applied for a patent on the cND technology through Harvard University (application number 62/363,395). If the patent is granted and commercial companies license the rights to the technology, the authors may receive royalty payments. However, plasmids for the nanodisc scaffolding proteins will be provided upon request. The authors declare no other competing financial interests.
Integrated supplementary information
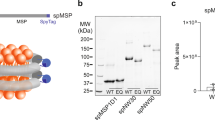
Supplementary Figure 1 Characterization of cNW9 by MS/MS confirms the ligation of the C terminus to the N terminus.
MS/MS spectrum of a tryptic fragment of cNW9 showing the ligation of the C-terminal motif (LNTQLPGTG-His6) to the N-terminal residues (GSTFSK). Expected masses for b and y ions along with the peptide sequence are listed in the table. The b and y ions that were identified in the MS/MS spectrum are highlighted in blue and red. The full amino acid sequence of linear NW9 is shown at the top.
Supplementary Figure 2 Characterization of cNW30 by MS/MS confirms the ligation of the C terminus to the N terminus.
MS/MS spectrum of a tryptic fragment of cNW30 showing the ligation of the C-terminal motif (LNTQLPGTG-His6) to the N-terminal residues (GSTFSK). Expected masses for b and y ions along with the peptide sequence are listed in the table. The b and y ions that were identified in the MS/MS spectrum are highlighted in blue and red. The full amino acid sequence of linear NW30 is shown at the top.
Supplementary Figure 3 Characterization of cNW50 by MS/MS confirms the ligation of the C terminus to the N terminus.
Spectrum of a tryptic fragment of cNW50 showing the ligation of the C-terminal motif (LNTQLPGTG-His6) to the N-terminal residues (GSTFSK). Expected masses for b and y ions along with the peptide sequence are listed in the table. The b and y ions that were positively identified in the MS/MS spectrum are highlighted in blue and red. The amino acid sequence of linear NW50 (after TEV cleavage) is shown at the top.
Supplementary Figure 4 Diameter distribution for nanodiscs assembled using cNW30, NW30 and cNW50 proteins.
(a) Diameter distribution for nanodiscs assembled using cNW30 (left) and NW30 (right). There is less variance in the lengths of cNW30 nanodiscs compared to NW30 nanodiscs (p=0.002). (b) Diameter distribution for nanodiscs made using cNW50. In the box-and-whisker plots, center lines show the means; box limits indicate the 25th and 75th percentiles; whiskers extend to the 5th and 95th percentiles. Raw data (jittered along x for clarity) are shown next to its representative plot. There is less variance in the lengths of cNW30 compared to NW30 nanodiscs (p = 0.002). Measurements were performed with the ImageJ software30.
Supplementary Figure 5 Producing covalently circularized nanodiscs with defined sizes and shapes.
(a) SDS-PAGE gel showing the final products after circularization over Ni beads with or without lipids. (b) Adding evolved sortase to diluted NW11 solution results mainly in NW11 circularization. Sortase was added to a dilute NW11 solution ([NW11] < 15 μM) to suppress linking two or more copies of NW11. (c) Adding evolved sortase to a concentrated NW11 solution leads to multimerization followed by circularization. (d) Negative-stain EM images showing large nanodiscs made using oligomeric circularized NW11. (e) Cryo-EM images of individual nanodiscs with different shapes. A triangle-shaped nanodisc is made using 3 copies of NW11 covalently linked together and circularized. Adding more lipids can change the shape back to a circular disc. Right: potential molecular arrangements of NW11 molecules around the differently shaped nanodiscs. (f) SDS-PAGE analysis of NW30 before and after circularization. Circularized NW30 migrates slower than its linear form. Right: negative-stain EM analysis of the nanodiscs made using circularized NW30 (cNW30) shows the formation of ~15 nm nanodiscs. (g) SDS-PAGE analysis of NW50 before and after circularization. Uncropped images are shown in Supplementary note 2. Right: negative-stain EM analysis of the nanodiscs made using circularized NW50 (cNW50) shows the formation of ~50 nm nanodiscs. Bottom: Negative-stain (left) and cryo-EM images (right) for individual nanodiscs showing top and side views. Scale bar, 50 nm.
NW30 and NW50 are sortaggable variants that we designed to assemble ~ 30- and 50-nm nanodiscs, respectively. Surprisingly, the cNW30 forms very homogenous ~15 nm instead of 30 nm nanodiscs On the other hand, as predicted, circularized NW50 assembled mainly ~ 50 nm nanodiscs.
Supplementary Figure 6 The oligomeric, circularized species containing variable numbers of NW11 assemble into nanodiscs of various sizes.
Adding evolved sortase to concentrated NW11 solution leads to multimerization of NW11 followed by circularization. Bottom: Negative-stain EM image showing large nanodiscs made using oligomeric, circularized NW11. Scale bar, 100 nm.
Supplementary Figure 7 Analysis of nanodiscs assembled using a cNW30:lipid ratio of 1:1000.
(a) Size-exclusion chromatography (SEC) analysis of the assembled nanodiscs. The SEC column (Superose 6 10/300) was equilibrated in 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA. The peak (labeled with *) was collected and analyzed by negative-stain EM (b). Scale bars, 15 nm (a) and 100 nm (b)
Supplementary Figure 8 Relaxation analysis of DMPC/DMPG (3:1) phospholipids in cNW and ΔH5 nanodiscs.
(a) 1H-NMR T1 relaxation times of DMPC/DMPG lipid signals in empty ΔH5 (blue) and cNW9 (red) nanodiscs. (b) 1H-NMR T2 relaxation times of DMPC/DMPG lipids signals in empty ΔH5 (blue) and cNW9 (red) nanodiscs (c) Chemical structure of DMPC and DMPG.
Except the DMPC γ methyl group located outside the bilayer region, the lipids in cNW9 show shorter T1 and T2 times consistent with smaller nanodisc size and dynamics indicative of the more restrictive circularized cNW9 belt. Measurements were acquired at 45ºC on a Bruker 500-MHz spectrometer using inversion recovery for T1 and CPMG refocusing train for T2.
Supplementary Figure 9 Covalent circularization enhances thermal stability of MSPs and embedded VDAC-1.
Thermal unfolding of MSP1D1 (black) and cNW11 (red) without (a) and with lipids (b) followed by circular dichroism (CD) spectroscopy at 222 nm, the wavelength most characteristic of helical secondary structure. A lipid mixture of POPC:POPG at a molar ratio of 3:2 was used to generate nanodiscs. (c) Reconstitution into MSP1D1 nanodiscs increases the melting temperature, Tm, of VDAC1 by 9.2°C over that of VDAC1 in an LDAO micelle environment, and covalent circularization of the scaffold protein (cNW11) raises the Tm by an additional 9.2°C. Thermal unfolding of human VDAC-1 was followed by CD spectroscopy at 218 nm, the wavelength most characteristic of β-sheet secondary structure. Orange: VDAC1 in 0.1% LDAO, black: VDAC1 reconstituted into conventional nanodiscs (assembled using MSP1D1), and red: VDAC1 reconstituted into circularized nanodiscs (assembled using cNW11). Nanodiscs were made with a 3:2 POPC:POPG mixture. (d) Thermal unfolding of VDAC1 reconstituted into circularized nanodiscs (assembled using cNW9) followed by CD spectroscopy at 218 nm. Nanodiscs were made with POPC/POPG lipids at a molar ratio of 3:2. All samples were in 20 mM Tris-HCl, pH 7.5, 100 mM NaCl. (e) Analysis of the VDAC1 nanodisc assembly reaction. Top: size-exclusion chromatography and negative-stain EM of VDAC1 in cNW9 nanodiscs. The negative-stain EM shows nanodiscs containing a single channel. The stain-filled channels appear as dark spots inside the nanodiscs. Bottom: SDS-PAGE analysis of the nanodisc assembly. Fractions 1-6 were collected and analyzed.
Supplementary Figure 10 Covalent circularization stabilizes MSPs against digestion by V8 protease without and with lipids.
(a, b) SDS-PAGE analysis of the proteolysis of lipid-free MSPΔH5 and cNW9. Samples were treated with V8 protease for 0 min (before addition of V8), and for 1, 3 and 18 hours. Lanes are labeled with times of protease treatment. Proteolysis was performed in 20 mM Tris-HCl, pH 7.5, 100 mM NaCl at 37°C and using a protein:protease ratio (w/w) of 1000:1. Treatment of MSPΔH5 with V8 resulted in the appearance of a large peptide, which is close in size to MSPΔH5 and is not discernible until about 18 hours after addition of V8; this peptide may be generated earlier but is not visible due to the low amount or overlap with uncleaved MSPΔH5. The intensity of the MSPΔH5 band decreased by 75% after 3 hours and by 93% after 18 hours. On the other hand, the intensity of the cNW9 band decreased by only 20% after 18 hours. A band that corresponds to linearized NW9 was observed after 3 hours and increased in intensity after 18 hours. (c, d) SDS-PAGE analysis of the proteolysis of nanodiscs assembled with MSPΔH5 and cNW9. Samples were treated with V8 protease for 0 min (before addition of V8), and for 20 min, 1, 3 and 18 hours. Proteolysis was performed at 37°C at pH 7.5 in 20 mM Tris-HCl, 100 mM NaCl using a 100:1 protein:protease (w/w). The band intensity of MSPΔH5 decreased by 81% after 3 hours. There was no decrease in the cNW9 band intensity up to 3 hours. The ImageJ software was used for analyzing band intensities.
Supplementary Figure 11 Single-particle EM analysis of negatively stained VDAC-1 in cNW9 nanodiscs.
(a) Representative EM image of VDAC-1 in cNW9. Right. Enlarged image of the rectangular area in panel a, showing one VDAC-1 channel per nanodisc. Images were collected on a Tecnai T12 electron microscope (FEI) operated at 120 kV. (b) Representative two-dimensional (2D) class averages of VDAC-1 in cNW9 nanodisc, showing distinct views (top, tilted and side views). (c) Cartoon representation of VDAC-1 in a cNW9 nanodisc. Scale bars, 100 nm (a) and 20 nm (b)
Supplementary Figure 12 Comparison of the quality of NMR spectra of VDAC-1 in ΔH5 and cNW9 nanodiscs.
(a) 1H-15N TROSY HSQC recorded at 45°C of 100 uM 15N-2D-labeled VDAC-1 in ΔH5 3:1 DMPC:DMPG nanodiscs acquired overnight on a 600-MHz spectrometer. (b) 1H-15N TROSY HSQC recorded at 45°C of 100 μM 15N-2D-labeled VDAC1 in cNW9 3:1 DMPC:DMPG nanodiscs acquired overnight on a 800-MHz spectrometer. The spectral quality and sample stability are greatly improved by using cNW9 as compared to ΔH5 nanodiscs.
Supplementary Figure 13 Cryo-EM images of poliovirus plus CD155-decorated 50-nm nanodiscs.
(a) Representative cryo-EM images of poliovirus plus CD155-decorated 50-nm nanodiscs in vitreous ice. Scale bar represents 100 nm. (b) Representative cryo-EM images of poliovirus-nanodisc complexes showing different intermediate states. Scale bars, 100 nm (a) and 50 nm (b).
We find it unlikely that the putative pore in the nanodisc is a hexametric arrangement of CD155 since we used only the ectodomains of the receptor linked to lipid molecules via His tags. Scale bars, 100 nm (a) and 50 nm (b)
Supplementary Figure 14 Cryo-EM images of poliovirus plus CD155-decorated 15-nm nanodiscs.
(a) Representative cryo-EM images of poliovirus plus CD155-decorated 15-nm nanodiscs in vitreous ice. Scale bar represents 100 nm. (b) Cryo-EM image showing individual viruses ejecting RNA after incubation with CD155-decorated 15-nm nanodiscs. Scale bar, 100 nm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Table 1, Supplementary Notes 1 and 2, and Supplementary Protocol. (PDF 4189 kb)
Rights and permissions
About this article
Cite this article
Nasr, M., Baptista, D., Strauss, M. et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods 14, 49–52 (2017). https://doi.org/10.1038/nmeth.4079
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4079
This article is cited by
-
Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel
Nature Communications (2024)
-
A novel strategy for partial purification of alkane hydroxylase from P. chrysogenum SNP5 through reconstituting its native membrane into liposome
Scientific Reports (2024)
-
Structural basis for the binding of DNP and purine nucleotides onto UCP1
Nature (2023)
-
Anti-nanodisc antibodies specifically capture nanodiscs and facilitate molecular interaction kinetics studies for membrane protein
Scientific Reports (2023)
-
Self-cyclisation as a general and efficient platform for peptide and protein macrocyclisation
Communications Chemistry (2023)