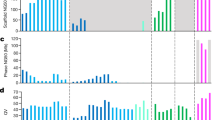
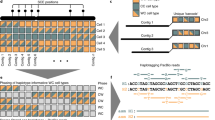
Abstract
While genome assembly projects have been successful in many haploid and inbred species, the assembly of noninbred or rearranged heterozygous genomes remains a major challenge. To address this challenge, we introduce the open-source FALCON and FALCON-Unzip algorithms (https://github.com/PacificBiosciences/FALCON/) to assemble long-read sequencing data into highly accurate, contiguous, and correctly phased diploid genomes. We generate new reference sequences for heterozygous samples including an F1 hybrid of Arabidopsis thaliana, the widely cultivated Vitis vinifera cv. Cabernet Sauvignon, and the coral fungus Clavicorona pyxidata, samples that have challenged short-read assembly approaches. The FALCON-based assemblies are substantially more contiguous and complete than alternate short- or long-read approaches. The phased diploid assembly enabled the study of haplotype structure and heterozygosities between homologous chromosomes, including the identification of widespread heterozygous structural variation within coding sequences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Goffeau, A. et al. Life with 6000 genes. Science 274, 546, 563–567 (1996).
Myers, E.W. et al. A whole-genome assembly of Drosophila. Science 287, 2196–2204 (2000).
Bonfield, J.K., Smith, Kf. & Staden, R. A new DNA sequence assembly program. Nucleic Acids Res. 23, 4992–4999 (1995).
Mouse ENCODE Consortium. et al. An encyclopedia of mouse DNA elements (mouse ENCODE). Genome Biol. 13, 418 (2012).
Celniker, S.E. et al. Unlocking the secrets of the genome. Nature 459, 927–930 (2009).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Earl, D. et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 21, 2224–2241 (2011).
Church, D.M. et al. Extending reference assembly models. Genome Biol. 16, 13 (2015).
Tewhey, R., Bansal, V., Torkamani, A., Topol, E.J. & Schork, N.J. The importance of phase information for human genomics. Nat. Rev. Genet. 12, 215–223 (2011).
Henson, J., Tischler, G. & Ning, Z. Next-generation sequencing and large genome assemblies. Pharmacogenomics 13, 901–915 (2012).
Alkan, C., Sajjadian, S. & Eichler, E.E. Limitations of next-generation genome sequence assembly. Nat. Methods 8, 61–65 (2011).
Vinson, J.P. et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 15, 1127–1135 (2005).
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007).
Iqbal, Z., Caccamo, M., Turner, I., Flicek, P. & McVean, G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat. Genet. 44, 226–232 (2012).
Kajitani, R. et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24, 1384–1395 (2014).
Roach, J.C. et al. Chromosomal haplotypes by genetic phasing of human families. Am. J. Hum. Genet. 89, 382–397 (2011).
Kirkness, E.F. et al. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 23, 826–832 (2013).
Kitzman, J.O. et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat. Biotechnol. 29, 59–63 (2011).
McCoy, R.C. et al. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PloS One 9, e106689 (2014).
Mostovoy, Y. et al. A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods 13, 587–590 (2016).
Berlin, K. et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33, 623–630 (2015).
Gordon, D. et al. Long-read sequence assembly of the gorilla genome. Science 352, aae0344 (2016).
Chin, C.-S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Fasulo, D., Halpern, A., Dew, I. & Mobarry, C. Efficiently detecting polymorphisms during the fragment assembly process. Bioinformatics 18, S294–S302 (2002).
The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Gan, X. et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477, 419–423 (2011).
Simão, F.A., Waterhouse, R.M., Ioannidis, P., Kriventseva, E.V. & Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Koren, S., Walenz, B.P., Berlin, K., Miller, J.R. & Phillippy, A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Preprint at bioRxiv http://dx.doi.org/10.1101/071282 (2016).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18 (2012).
Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19, ii215–ii225 (2003).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Patel, S., Swaminathan, P., Fennell, A. & Zeng, E. in Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (eds. Huan, J. et al.) 1771–1773 (EEE, 2015).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at arXiv:1207.3907v2 [q-bio.GN] (2012).
Bansal, V. & Bafna, V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics 24, i153–i159 (2008).
Degner, J.F. et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25, 3207–3212 (2009).
Liu, Y.-G. & Whittier, R.F. Rapid preparation of megabase plant DNA from nuclei in agarose plugs and microbeads. Nucleic Acids Res. 22, 2168–2169 (1994).
Hayward, G.S. Unique double-stranded fragments of bacteriophage T5 DNA resulting from preferential shear-induced breakage at nicks. Proc. Natl. Acad. Sci. USA 71, 2108–2112 (1974).
Myers, G. Algorithms in Bioinformatics (eds. Brown, D. & Morgenstern, B.) 52–67 (Springer, 2014).
Myers, E.W. The fragment assembly string graph. Bioinformatics 21, ii79–ii85 (2005).
Chaisson, M.J. & Tesler, G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238 (2012).
Acknowledgements
The sequencing of the Cabernet Sauvignon genome was supported in part by a gift from the J. Lohr Vineyards and Wines to D.C. We would also like to thank F. Neto for providing an early-release BUSCO plant data set. Clavicorona pyxidata DNA was provided by L. Nagy (Institute of Biochemistry Biological Research Centre of the Hungarian Academy of Sciences). We thank J. Puglisi, F. Jupe, A. Copeland, and A. Wenger for reading and critiquing the manuscript. The project was supported in part by National Institutes of Health award (R01-HG006677 to M.C.S.) and by National Science Foundation awards (DBI-1350041 and IOS-1237880 to M.C.S.; MCB 0929402; and MCB 1122246 to J.R.E.). J.R.E. is an investigator at the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (GBMF 3034).
Author information
Authors and Affiliations
Contributions
C-S.C., P.P., A.C., D.R.R., and M.C.S. conceived the idea of the FALCON–FALCON-Unzip assembler. C.-S.C, P.P., F.J.S., M.N., G.T.C., D.R.R., D.C., and M.C.S. designed the experiments and performed the analysis. P.P., D.C., D.R.R., and M.C.S. collected the sequencing data. R.O'M. C.L., and J.R.E. constructed the Col-0-Cvi-1. A.C., R.O'M. R.F.-B., A.M.-C., G.R.C., M.D., C.L., J.R.E., and D.C. collected the samples and prepared DNA for sequencing. C.-S.C., P.P., F.J.S., M.N., G.T.C., D.C., D.R.R., and M.C.S. wrote the manuscript. C.-S.C. and C.D. implemented the computer code.
Corresponding authors
Ethics declarations
Competing interests
C.-S.C., P.P., G.T.C., C.D., and D.R. are employees and shareholders of Pacific Biosciences, a company commercializing DNA sequencing technology.
Integrated supplementary information
Supplementary Figure 1 Schematics of the software and data process modules and the FACLON-Unzip assembly graph process for resolving haplotypes.
(a) Data dependence flow and software modules inside FALCON and FALCON-Unzip
(b) Left: Initial assembly graph of a contig in the Arabidopsis F1 hybrid assembly. The different colors represent different haplotype blocks and phases. Right: The assembly graph after “unzipping”. Conceptually, the unzipping step identifies the heterozygous SNPs and uses them to remove overlaps between reads from different haplotypes. After removing such overlaps, nodes from the different haplotypes in the assembly graph will no longer have edges between them. This allows FALCON-Unzip to identify long haplotype specific paths and construct haplotigs of them. The dashed circle region indicates haplotype blocks that can be extended through a bubble region.
Supplementary Figure 4 Assemblytic analysis comparison of the Arabidopsis F1 assemblies from FALCON-Unzip, Platanus, and SOAPdenovo.
(a) Cumulative sequence length of three Arabidopsis F1 assemblies created by FALCON-Unzip, Platanus, and SOAPdenovo compared to the TAIR10 reference. (b) Variants called using Assemblytics from three Arabidopsis F1 assemblies created by FALCON-Unzip,Platanus, and SOAPdenovo.
Supplementary Figure 7 Assembly comparison: FALCON-Unzip V. vinifera cv. Cabernet Sauvignon assembly versus V. vinifera reference genome
(a) MUMmerplot of FALCON-Unzip V. vinifera cv. Cabernet Sauvignon assembly versus V. vinifera reference genome. For clarity only alignments >= 10,000 bp long to the primary chromosomes are displayed. (b) The synteny between PN40024 Chr1 from 5’- telomere to centromere (green line) to the longest contig 000000F (black line) and its associated haplotigs (blue lines). The vertical green and blue lines indicated homologous coding sequences between the sequences. The cyan lines in the bottom indicate the synteny between the primary contig and other primary contigs. (c) Synteny alignment between two primary contigs 000334F vs. 000000F. (d) Synteny alignment between two primary contigs 000057F vs 000075F.
Supplementary Figure 8 Comparison of the distribution the het-SNP site density of the three genomes
(a) The distribution of number of het-SNPs observed of the reads used for phasing of the longest contig of each genome in semi-log plot. (b) Fitting the distributions with a exponential function (density ~ c * exp(-a * het-SNP count)). We pick het-SNP count range of 10 to 200 for Arabidopsis, 50 to 200 for Vitis, and 10 to 100 for Clavicorona to catch the exponential decay part. The fitted parameter a = -0.0222, 0.0216, 0.0412 for Arabidopsis, Vitis and Clavicorona respectively. The fastest decay rate for Clavicorona indicates it has the least variation between the haplotypes among the three genomes. From this fitting, we expect to see about 45 (Arabidopsis), 46 (Vitis), and 24 (Clavicorona) per 10kb in the regions of interests.
Supplementary Figure 9 Example of a low heterozygosity region observed in Clavicorona genome.
The het-SNPs are called with FreeBayes on the alignments of the short read data to only the primary contigs. The contig 00003F has a low heterozygosity region from ~1.2Mb to ~2.7Mb.
Supplementary Figure 11 Candidates for differentially expressed alleles from RNA-seq data.
(a)(b)We mapped both genomic reads (middle panel) and cDNA reads (lower panel) to the primary contigs from our Clavicorona pyxidata assembly. We also shows curated CDS sequences mapped to the contig (top panel). The genomic reads shows both alleles mapped while we only observe on major allele in the transcript reads.
Supplementary Figure 15 Summary of the greedy SNP phasing algorithm.
(a) All pairs of het-SNPs that are covered by multiple reads are evaluation. A “coupling score” is calculation from the number reads that support current haplotype assignment of the het-SNPs. (b)(c) We linearly scan through the het-SNP positions. If the total score is improved by flipping the haplotype assigned at one location, then we flip the assignment. (d) An example showing the “coupling score” before the flipping process (un-phased het-SNPs assignment) and afterward (phased het-SNP assignment).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1–10 and Supplementary Note 1 (PDF 4833 kb)
Supplementary Data 1
SNP identified by nucmer between FALCON col-0 assembly and the TAIR10 reference (TXT 1920 kb)
Supplementary Data 2
List of syntenic regions identify between different primary contigs of the FALCON-Unzip Arabidopsis thaliana Col-0 x Cvi-1 assembly. (CSV 21668 kb)
Supplementary Data 3
List of syntenic regions identify between different primary contigs of the FALCON-Unzip Vitis vinifera assembly. (CSV 2183 kb)
Supplementary Data 4
Example of starting an AWS instance to run FALCON/FALCON-Unzip for Clavicorona pyxidata assembly (PDF 2523 kb)
Rights and permissions
About this article
Cite this article
Chin, CS., Peluso, P., Sedlazeck, F. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13, 1050–1054 (2016). https://doi.org/10.1038/nmeth.4035
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4035
This article is cited by
-
Whole-genome sequencing and analysis of Chryseobacterium arthrosphaerae from Rana nigromaculata
BMC Microbiology (2024)
-
De novo Phased Genome Assembly, Annotation and Population Genotyping of Alectoris Chukar
Scientific Data (2024)
-
Chromosome-level genome assembly of Platycarya strobilacea
Scientific Data (2024)
-
Transcriptional effects of carbon and nitrogen starvation on Ganoderma boninense, an oil palm phytopathogen
Molecular Biology Reports (2024)
-
Genomic insights into biased allele loss and increased gene numbers after genome duplication in autotetraploid Cyclocarya paliurus
BMC Biology (2023)