Abstract
Advances in high-resolution cryo-electron microscopy (cryo-EM) require the development of validation metrics to independently assess map quality and model geometry. We report EMRinger, a tool that assesses the precise fitting of an atomic model into the map during refinement and shows how radiation damage alters scattering from negatively charged amino acids. EMRinger (https://github.com/fraser-lab/EMRinger) will be useful for monitoring progress in resolving and modeling high-resolution features in cryo-EM.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Liao, M., Cao, E., Julius, D. & Cheng, Y. Curr. Opin. Struct. Biol. 27, 1–7 (2014).
Shi, Y. Cell 159, 995–1014 (2014).
Wong, W. et al. eLife 3, e03080 (2014).
Henderson, R. et al. Structure 20, 205–214 (2012).
Scheres, S.H. & Chen, S. Nat. Methods 9, 853–854 (2012).
Chen, V.B. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Ramachandran, G.N., Ramakrishnan, C. & Sasisekharan, V. J. Mol. Biol. 7, 95–99 (1963).
Lovell, S.C., Word, J.M., Richardson, J.S. & Richardson, D.C. Proteins 40, 389–408 (2000).
DiMaio, F., Zhang, J., Chiu, W. & Baker, D. Protein Sci. 22, 865–868 (2013).
Amunts, A. et al. Science 343, 1485–1489 (2014).
Falkner, B. & Schroder, G.F. Proc. Natl. Acad. Sci. USA 110, 8930–8935 (2013).
Brown, A. et al. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015).
Lang, P.T., Holton, J.M., Fraser, J.S. & Alber, T. Proc. Natl. Acad. Sci. USA 111, 237–242 (2014).
Lang, P.T. et al. Protein Sci. 19, 1420–1431 (2010).
Zhou, A.Q., O'Hern, C.S. & Regan, L. Proteins 82, 2574–2584 (2014).
Shapovalov, M.V. & Dunbrack, R.L. Jr. Structure 19, 844–858 (2011).
Dunbrack, R.L. Jr. Curr. Opin. Struct. Biol. 12, 431–440 (2002).
Liao, M., Cao, E., Julius, D. & Cheng, Y. Nature 504, 107–112 (2013).
DiMaio, F. et al. Nat. Methods 12, 361–365 (2015).
Allegretti, M., Mills, D.J., McMullan, G., Kuhlbrandt, W. & Vonck, J. eLife 3, e01963 (2014).
Li, X. et al. Nat. Methods 10, 584–590 (2013).
Bartesaghi, A., Matthies, D., Banerjee, S., Merk, A. & Subramaniam, S. Proc. Natl. Acad. Sci. USA 111, 11709–11714 (2014).
Fioravanti, E., Vellieux, F.M., Amara, P., Madern, D. & Weik, M. J. Synchrotron Radiat. 14, 84–91 (2007).
Mitsuoka, K. et al. J. Mol. Biol. 286, 861–882 (1999).
Fernández, I.S. et al. Science 342, 1240585 (2013).
Havranek, J.J. & Baker, D. Protein Sci. 18, 1293–1305 (2009).
Cowtan, K. IUCrJ 1, 387–392 (2014).
Terwilliger, T.C. Acta Crystallogr. D Biol. Crystallogr. 66, 268–275 (2010).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Acknowledgements
This work benefited from helpful discussions with D. Agard, D. Baker, E. Green, C. Greenberg, A. Frost and S. Scheres. B.A.B. is supported by US National Institutes of Health (NIH) training grant T32GM008284. Y.C. is supported by US NIH grants GM082893, GM098672 and GM082250. N.E. and P.D.A. are supported by US NIH grant GM063210, the Phenix Industrial Consortium and, in part, by the US Department of Energy under contract DE-AC02-05CH11231. J.S.F. is supported by a Searle Scholar award from the Kinship Foundation, a Pew Scholar award from the Pew Charitable Trusts, a Packard Fellow award from the Lucille and David Packard Foundation, US NIH grants OD009180 and GM110580, US National Science Foundation grant STC-1231306 and a UCSF-SABRE Innovation grant.
Author information
Authors and Affiliations
Contributions
B.A.B., N.E., P.D.A. and J.S.F. designed research. B.A.B. and N.E. wrote the EMRinger code. R.Y.-R.W. and F.D. refined models. Y.C. contributed data sets. B.A.B. and J.S.F. wrote the manuscript. All authors commented on and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
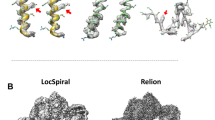
Supplementary Figure 1 Atomic models in cryo-EM maps.
(a) Two alpha (blue) and two beta (green) subunits of the T20S proteasome are shown as cartoon tubes fitted in a 3.2 Å potential map at isolevel 0.25 (EMDB 5623, PDB 3J9I). (b) The same subunits are depicted in density at a higher isolevel of 0.35, where sharper features of side chain density can be observed. (c) Two subunits of the TrpV1 tetramer are shown in green and blue in a 3.27Å potential map at an isolevel of 10 (EMDB 5778, PDB 3J9J)
Supplementary Figure 2 EMRinger score is unaffected by model size.
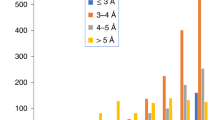
(a) EMRinger plot for a 366 amino acid monomer of the Hepatitis B virus capsid gives a peak EMRinger score of 3.25 (EMDB 2278, PDB 3J2V). (b) Histogram of EMRinger map value peaks above threshold 6.090 (the threshold of maximum EMRinger score) for the monomer in density. (c) EMRinger plot for the full biological 21960 amino acid 60-mer assembly of the Hepatitis B capsid gives a nearly identical set of scores to the monomer, with a peak score of 3.16. The smoother plot is likely due to the averaging out of artifacts due to grid sampling. (d) Histogram of EMRinger map value peaks above threshold 5.726 (the threshold of maximum EMRinger score) for the 60-mer in density.
Supplementary Figure 4 Controls to demonstrate how EMRinger score is affected by low-pass filtering, changes to grid spacing and adjustment of the sampling frequency.
(a) EMRinger Score degrades rapidly with decreasing resolution. The T20S proteasome map (EMDB 5623, PDB 3J9I) is low-pass filtered to resolutions ranging from 3.2 to 7 Å. EMRinger scores for each of these filtered maps show a resolution dependence and that by 5 Å resolution side chains are no longer distinguishable from noise and the EMRinger score is near 0. (b) EMRinger scores are largely robust to changes in grid spacing. The T20S proteasome map is modified to have a range of adjusted grid spacing, and EMRinger score is calculated for each map against the deposited models. The EMRinger score decreases by a maximum of 17% with a maximum increase of grid spacing before lowering resolution due to the Nyquist limit. (c) EMRinger score decreases with increased sampling angle. The EMRinger score is calculated with different sampling angles from 1° to 15°, with a null hypothesis based on the correct number of bins that are rotameric for each sampling angle and with a standard fixed null hypothesis of 50%. This shows that EMRinger score decreases with Sampling Angle as a result of the changes in null hypothesis rather than significant differences in enrichment. (d) Interpretation of EMRinger score is not affected by choice of standard sampling angle of 5°. EMRinger scores were calculated for structures from table S1 at sampling angles of 1° and 5° and the results were compared. The linear correlation between the two (R2=0.98) sampling angles indicates that the comparison of scores will be the same regardless of sampling angle, so long as a single sampling angle is used for comparison.
Supplementary Figure 5 Histograms of TrpV1 models at multiple map value thresholds.
(a) Histograms at thresholds of 4, 8, 12, and 16 for EMRinger map value peaks of the transmembrane region of the deposited TrpV1 model (EMDB 5778, PDB 3J5P). (b) Histograms at thresholds of 4, 8, 12, and 16 for the EMRinger map value peaks of the transmembrane region of TrpV1 refined by Rosetta refinement show improved enrichment at rotameric positions at all thresholds.
Supplementary Figure 6 EMRinger scores report on effective refinement of atomic models into EM maps.
(a) The EMRinger score improves during refinement. Rosetta refinement trajectories for 9 trials are shown in light green with the final refinement shown in dark green. (b) Map value cutoff scan for the unrefined model of TrpV1 (red, EMDB 5778, PDB 3J5P), the transmembrane region of the deposited TrpV1 model (orange), and for the model of TrpV1 refined by Rosetta (green, PDB 3J9J) show the improvement during refinement. (c) Analyzing the unrefined (red) and refined (green) models in the transmembrane region highlights how portions of the model experience dramatic increases in rotameric peaks after refinement. (d) The unrefined (red) and refined (green) TrpV1 models are shown in density (isolevel of 10), revealing that small shifts in the placement of backbone of the alpha helix improves EMRinger statistics.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Table 1 (PDF 1195 kb)
Supplementary Software
Phenix Scripts: Scripts to calculate EMRinger scores, real-space cross-correlation scores, and rolling window metrics, using phenix.python. (TAR 42 kb)
Rights and permissions
About this article
Cite this article
Barad, B., Echols, N., Wang, RR. et al. EMRinger: side chain–directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946 (2015). https://doi.org/10.1038/nmeth.3541
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3541
This article is cited by
-
Mechanism of anion exchange and small-molecule inhibition of pendrin
Nature Communications (2024)
-
The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center
Nature Chemical Biology (2024)
-
Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs
Nature Communications (2023)
-
Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir
Nature Communications (2023)
-
Structural insights into the broad protection against H1 influenza viruses by a computationally optimized hemagglutinin vaccine
Communications Biology (2023)