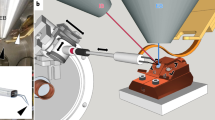
Abstract
We developed a method for visualizing tissues from multicellular organisms using cryo-electron tomography. Our protocol involves vitrifying samples with high-pressure freezing, thinning them with cryo-FIB-SEM (focused-ion-beam scanning electron microscopy) and applying fiducial gold markers under cryogenic conditions to the lamellae post-milling. We applied this protocol to acquire tomograms of vitrified Caenorhabditis elegans embryos and worms, which showed the intracellular organization of selected tissues at particular developmental stages in otherwise intact specimens.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Fridman, K., Mader, A., Zwerger, M., Elia, N. & Medalia, O. Nat. Rev. Mol. Cell Biol. 13, 736–742 (2012).
Yahav, T., Maimon, T., Grossman, E., Dahan, I. & Medalia, O. Curr. Opin. Struct. Biol. 21, 670–677 (2011).
Gan, L. & Jensen, G.J. Q. Rev. Biophys. 45, 27–56 (2012).
Luč ič, V., Rigort, A. & Baumeister, W. J. Cell Biol. 202, 407–419 (2013).
Dobro, M.J., Melanson, L.A., Jensen, G.J. & McDowall, A.W. Methods Enzymol. 481, 63–82 (2010).
Dubochet, J. & Sartori Blanc, N. Micron 32, 91–99 (2001).
Dubochet, J. et al. Q. Rev. Biophys. 21, 129–228 (1988).
Marko, M., Hsieh, C., Schalek, R., Frank, J. & Mannella, C. Nat. Methods 4, 215–217 (2007).
Rigort, A. et al. Proc. Natl. Acad. Sci. USA 109, 4449–4454 (2012).
Wang, K., Strunk, K., Zhao, G., Gray, J.L. & Zhang, P. J. Struct. Biol. 180, 318–326 (2012).
Hsieh, C., Schmelzer, T., Kishchenko, G., Wagenknecht, T. & Marko, M. J. Struct. Biol. 185, 32–41 (2014).
Grimm, R., Typke, D., Barmann, M. & Baumeister, W. Ultramicroscopy 63, 169–179 (1996).
Dahl, R. & Staehelin, L.A. J. Electron Microsc. Tech. 13, 165–174 (1989).
Burke, B. & Stewart, C.L. Nat. Rev. Mol. Cell Biol. 14, 13–24 (2013).
Ho, C.Y. & Lammerding, J. J. Cell Sci. 125, 2087–2093 (2012).
Ben-Harush, K. et al. J. Mol. Biol. 386, 1392–1402 (2009).
Grossman, E. et al. J. Struct. Biol. 177, 113–118 (2012).
Kipreos, E.T. Nat. Rev. Mol. Cell Biol. 6, 766–776 (2005).
Strange, K., Christensen, M. & Morrison, R. Nat. Protoc. 2, 1003–1012 (2007).
Hayles, M.F., Stokes, D.J., Phifer, D. & Findlay, K.C. J. Microsc. 226, 263–269 (2007).
Hiramatsu, H. & Osterloh, F.E. Chem. Mater. 16, 2509–2511 (2004).
Gruska, M., Medalia, O., Baumeister, W. & Leis, A. J. Struct. Biol. 161, 384–392 (2008).
Nickell, S. et al. J. Struct. Biol. 149, 227–234 (2005).
Acknowledgements
We thank Y. Gruenbaum (The Hebrew University of Jerusalem) for providing us with the strains of C. elegans used in this study. This work was supported by a European Research Council (ERC) Starting Grant (243047 INCEL) and the Swiss National Science Foundation (SNSF 31003A_141083/1).
Author information
Authors and Affiliations
Contributions
J.H., A.K., K.T.S., D.B. and O.M. designed the experiments and wrote the manuscript. J.H. and M.B. carried out the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Cryo-holder design and overview of the cryo-FIB-SEM procedure carried out on embryos and worms
(a) The prototype cryo-holder. The attachment slit (black arrow) and shutter (black asterisk) were redesigned to accommodate standard EM grids at a 6° angle with respect to the FIB column in the Auriga crossbeam system. (b) The two-piece attachment slit is regulated by a screw (black arrow head) on the upper component (red arrow head), while the base is freely movable (green arrow head). (c) The aluminum shutter envelops the cryo-holder while attached to the tip of the rod of the VCT100 shuttle via a Teflon ring. (d) The cryo-holder inside the Auriga. (e) SEM image of a well-vitrified embryo. The lamella is flat and no cracks are present. (f) SEM image of a cracked embryo. Cracks in the sample (white arrow heads) can be observed immediately upon the removal of the top of the sample with the cryo-FIB. (g) Cryo-TEM image of an embryo lamella showing the full dimensions of a lamella used for tomographic imaging. White asterisk indicates areas used for focusing and tracking and white arrows show the direction of the Ga+ beam milling. (h) SEM image of a vitrified worm (white arrows) after the sublimation of 2-methylpentane. (i) FIB image of the worm lamella (area in between white arrow heads) after milling. (j) SEM image of the worm lamella from top showing the full dimensions of the milled area (white double arrow) and the non-milled sides (black double arrows). Scale bars represent 2 µm (g), 5 µm (e,i), 10 µm (f,j), and 100 µm (h).
Supplementary Figure 2 Cryo-electron microscopy of gold nanoparticles and CET of vitrified C. elegans worms
(a) Cryo-TEM image of gold nanoparticles synthesized in toluene and spread on the surface of a plunge frozen EM grid. The grid is covered by ice contamination that varies both in thickness and size (white asterisk) and shows the markers clustering within this contamination (black arrows). (b) Cryo-TEM image of gold nanoparticles synthesized in 2-methylpentane and spread on the surface of a plunge-frozen EM grid. Individual markers are evenly distributed along the grid surface and contamination is barely detectable (compare the areas indicated by the white asterisks). (c,e) 3.4 nm thick tomographic slices acquired with a –16 µm defocus on a worm cryo-lamella. Green arrow heads point towards filamentous structures adjacent to the INM, the black arrow points to a plasma membrane protrusion and the white asterisks indicate the position of NPCs. (N – nucleus, C – Cytoplasm). (d) A surface rendered view corresponding to the black-framed area in panel a. Plasma membrane – dark blue, nuclear membrane – pink, ribosomes – gold, NPCs – light blue, and filamentous structures adjacent to the INM – green. Final resolution of both tomograms was estimated from the first zero of the CTF and calculated to be 5.4 nm. Scale bars represent 250 nm (a,b) and 200 nm (c,e).
Supplementary Figure 3 Correlative light and electron microscopy of C. elegans embryos expressing lamin-GFP
(a) SEM overview of the grid before milling. The embryo that will be milled is indicated by the white arrow. (b) Bright-field-fluorescence channel overlay showing the same embryo post-milling, fixed in 4% PFA (white arrow). (c) SEM image of the embryo post-milling. White asterisk indicated the area where platinum was deposited inside the cryo-FIB-SEM device to facilitate milling. (d) Bright-field-fluorescence channel overlay showing bright lamin-GFP signal inside the thicker, non-milled sides of the embryo, and its distribution throughout the thin lamella. The white asterisk indicated the area where platinum was deposited (same as in panel c). The bright-field-fluorescence images (b,d) are rotated approximately 40° clock-wise compared to the SEM images (a,c). Scale bars indicate 200 µm (a), 50 µm (b), 5 µm (c), and 10 µm (d).
Supplementary Figure 4 Cryo-FIB milling of Drosophila melanogaster embryos
(a) SEM overview image of the EM grid bearing 3 D. melanogaster embryos (red arrows). (b) FIB side view of the edge of the embryo after several rounds of milling with high currents. (c) FIB side view of the thin lamella after successive rounds of course and fine milling. The area underneath the lamella is also cleared in order to facilitate tomographic imaging. (d) SEM top view of the lamella after milling. Black double arrows indicate the non-milled slopes surrounding the lamella and the white double arrows indicate the lamella itself. Scale bars indicate 500 µm (a) and 50 µm (b–d).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 572 kb)
CET of C. elegans embryos
The video shows the full tomogram from figure 2, panel c. (MOV 8206 kb)
CET of C. elegans worms
The video show the full tomogram from supplementary figure 2, panel c. (MOV 7758 kb)
Rights and permissions
About this article
Cite this article
Harapin, J., Börmel, M., Sapra, K. et al. Structural analysis of multicellular organisms with cryo-electron tomography. Nat Methods 12, 634–636 (2015). https://doi.org/10.1038/nmeth.3401
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3401
This article is cited by
-
Waffle Method: A general and flexible approach for improving throughput in FIB-milling
Nature Communications (2022)
-
Zooming in on host-symbiont interactions: advances in cryo-EM sample processing methods and future application to symbiotic tissues
Symbiosis (2022)
-
Direct imaging of uncoated biological samples enables correlation of super-resolution and electron microscopy data
Scientific Reports (2018)
-
The molecular architecture of lamins in somatic cells
Nature (2017)
-
Biological application of Compressed Sensing Tomography in the Scanning Electron Microscope
Scientific Reports (2016)