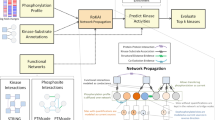
Abstract
Protein phosphorylation is important in cellular pathways and altered in disease. We developed MIMP (http://mimp.baderlab.org/), a machine learning method to predict the impact of missense single-nucleotide variants (SNVs) on kinase-substrate interactions. MIMP analyzes kinase sequence specificities and predicts whether SNVs disrupt existing phosphorylation sites or create new sites. This helps discover mutations that modify protein function by altering kinase networks and provides insight into disease biology and therapy development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Pawson, T. Nature 373, 573–580 (1995).
Reimand, J., Hui, S., Jain, S., Law, B. & Bader, G.D. FEBS Lett. 586, 2751–2763 (2012).
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanam, S. Science 298, 1912–1934 (2002).
Reimand, J. & Bader, G.D. Mol. Syst. Biol. 9, 637 (2013).
Reimand, J., Wagih, O. & Bader, G.D. PLoS Genet. 11, e1004919 (2015).
Riaño-Pachón, D.M. et al. BMC Genomics 11, 411 (2010).
Savas, S. & Ozcelik, H. BMC Cancer 5, 107 (2005).
Radivojac, P. et al. Bioinformatics 24, i241–i247 (2008).
Ren, J. et al. Mol. Cell. Proteomics 9, 623–634 (2010).
Ryu, G.M. et al. Nucleic Acids Res. 37, 1297–1307 (2009).
Reimand, J., Wagih, O. & Bader, G.D. Sci. Rep. 3, 2651 (2013).
Hornbeck, P.V. et al. Nucleic Acids Res. 40, D261–D270 (2012).
Diella, F. et al. BMC Bioinformatics 5, 79 (2004).
Keshava Prasad, T.S. et al. Nucleic Acids Res. 37, D767–D772 (2009).
Newman, R.H. et al. Mol. Syst. Biol. 9, 655 (2013).
Kel, A.E. et al. Nucleic Acids Res. 31, 3576–3579 (2003).
Fraley, C. & Raftery, A.E. J. Am. Stat. Assoc. 97, 611–631 (2002).
Weinstein, J.N. et al. Nat. Genet. 45, 1113–1120 (2013).
Aberle, H., Bauer, A., Stappert, J., Kispert, A. & Kemler, R. EMBO J. 16, 3797–3804 (1997).
Wu, L., Ma, C.A., Zhao, Y. & Jain, A. J. Biol. Chem. 286, 2236–2244 (2011).
Gully, C.P. et al. Proc. Natl. Acad. Sci. USA 109, E1513–E1522 (2012).
Gfeller, D., Ernst, A., Jarvik, N., Sidhu, S.S. & Bader, G.D. PLoS ONE 9, e94507 (2014).
Smyth, G.K. Stat. Appl. Genet. Mol. Biol. 3, Article3 (2004).
Magrane, M. Database 2011, bar009 (2011).
Croft, D. et al. Nucleic Acids Res. 39, D691–D697 (2011).
Ruepp, A. et al. Nucleic Acids Res. 38, D497–D501 (2010).
Reimand, J., Arak, T. & Vilo, J. Nucleic Acids Res. 39, W307–W315 (2011).
Merico, D., Isserlin, R. & Bader, G.D. Methods Mol. Biol. 781, 257–277 (2011).
Acknowledgements
We thank A. Moses for detailed comments that improved the method and the Kinexus Bioinformatics Corporation for conducting kinase assays. This work was supported by the Canadian Institutes of Health Research grant MOP-84324 to G.D.B.
Author information
Authors and Affiliations
Contributions
O.W., J.R. and G.D.B. devised the method and designed the analysis. O.W. analyzed the data, implemented the method and developed the software. O.W. wrote the initial manuscript. All authors edited and approved the final manuscript. J.R. and G.D.B. jointly supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Kinase-associated phosphorylation sites.
(left) Pie chart depicting proportions of tyrosine (white) and serine-threonine (black) kinases used in this study (right) The distribution of the number of phosphosites that are experimentally annotated to kinases.
Supplementary Figure 2 Iterative model refinement.
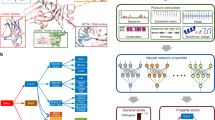
(a) Workflow of model refinement that discards sequences that do not correspond to the motif's general pattern. 1. An initial PWM is constructed using the positive set of kinase phosphorylation sites that is used to score the positive and negative sites. 2. The threshold t is defined as the score at the 90th percentile of the negative distribution of scores. 3. Positive sequences with a score below t are discarded. A new PWM is then constructed with the retained sequences. 4. This process is repeated until there are no further sequences to discard (i.e. all sequence achieved a score greater than t), or, when discarding sequences, result in a retained set of less than 10 sequences. (b) Examples of refinement for six kinases. Sequence logos show the underlying motif of kinase phosphorylation sites before and after the refinement procedure.
Supplementary Figure 3 Area under the curve (AUC) distribution of kinase specificity models.
(left) AUCs computed using kinase binding sites from all other kinase families as the negative set. AUCs below 0.6 (red line) were discarded from the analysis. (right) AUCs computed using a background of random unphosphorylated STY-centered sites. All retained models have AUCs greater than 0.64 (red line).
Supplementary Figure 4 Distribution of pSNVs across different positions in phosphosites.
(top) Bar plot shows the information content for each position, relative to the central residue. The information content was computed on all known kinase substrates. (bottom) Number of network-rewiring pSNVs relative to distance from phosphorylated residue. Significant proportions compared to non-rewiring pSNVs are marked with an asterisk (P<0.05, binomial test).
Supplementary Figure 5 Effect of number of kinase targets on called pSNVs.
Kinases with more experimentally validated targets likely have a larger number of called pSNVs. (a) shows the relationship between the number of experimentally validated kinase targets (after refinement, AUC≥0.6) vs. the number of pSNVs predicted to rewire the kinase. Correlations and their P-values are presented in the top left corner. The line of best fit is shown in red. (b) shows the same data represented in a box plot, showing a significant enrichment of the number of pSNVs for kinases with ≥100 phosphorylation targets. P-value represents enrichment as computed by a one-sided Wilcoxon signed-rank test.
Supplementary Figure 6 Pathway enrichment.
Pathway enrichment. Enrichment map showing pathways and processes with frequent network-rewiring mutations in phosphosites (FDR P<0.01, Poisson exact test). Edges connect pathways with many shared genes. Node size represents the number of rewiring mutations in the pathway.
Supplementary Figure 7 Colocalization and coexpression of rewired kinase-substrate pairs.
(a) pSNVs involved in rewiring (dark blue) are more likely to occur in unstructured regions compared to non-rewiring pSNVs (white). P-value above the bar reflects significantly higher number of pSNVs and is computed using one tailed binomial test (b) Expression and localization data were used to show rewired kinase- substrates (blue) are more likely to be co-expressed (left) and co-localized (right) compared to expectation from random kinase-substrate pairs (grey). P-values above bars were computed using the Z-test and represent a significantly higher number of rewiring pSNVs compared to randomly samples kinase-substrate pairs. Error bars represent the 95% confidence intervals.
Supplementary Figure 8 Experimental validation of kinase-substrate rewiring.
Nine experimentally validated (a) loss and (b) gain-of-phosphorylation events. Five network-rewiring mutations were selected as top ranking in terms of the patient sample count. For each of mutation, we selected top-ranking kinases rewired by that mutation in terms of the log ratio between wild type and mutant MSS scores. The bar plots quantify in vitro kinase activity in replicates of two for wild type and mutant peptide sequences as well as negative controls (blank). P-values represent the significance of difference between the wild type and mutant kinase activity, computed using an empirical Bayes moderated t-test and corrected for multiple testing using the Benjamini-Hochberg method of False Discovery Rate. The last four rewiring events were assayed against close family members of the rewired kinase instead of the exact kinase for further experimental support.
Supplementary Figure 9 Properties of samples containing TP53-R282W.
(a) Types of mutations in TP53 across samples with the TP53-R282W mutation. Only four of 23 samples show possibly deleterious mutations, such as frameshift deletions or nonsense mutations. (b) Samples with the rewiring mutation TP53-R282W show mRNA expression levels of TP53 that are similar to other samples. Samples with frameshift deletions (square) or nonsense mutations (triangle) are highlighted as points on the plot. One sample containing a frame shift mutation did not have a measured expression value for TP53. These two observations suggest that our predicted network-rewiring mutations in TP53 are active in corresponding cancer samples.
Supplementary Figure 10 Validation of TP53 expression in samples with the TP53-R282W mutation.
(a) Higher TP53 protein levels in samples containing mutations R213Q and R282W compared to other samples. These mutations are predicted to disrupt sites required for degradation or transcriptional repression of TP53 (b) Higher expression levels of LEF1, a downstream transcriptional target of CTNNB1, in samples containing mutations S37C and S37F. These mutations are predicted to disrupt phosphorylation in sites responsible for degradation of β-catenin. P-values in panels are based on a one-sided Wilcoxon signed-rank test.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Results, Supplementary Discussion, Supplementary Note and Supplementary Table 1 (PDF 1656 kb)
Supplementary Data 1
Kinase substrate data (ZIP 223 kb)
Supplementary Data 2
Negative phosphorylation data (TXT 312 kb)
Supplementary Data 3
Sequence logos (ZIP 1397 kb)
Supplementary Data 4
Mutation data (TXT 13800 kb)
Supplementary Data 5
Phosphorylation data (TXT 10806 kb)
Supplementary Data 6
TCGA network rewiring events (TXT 5931 kb)
Rights and permissions
About this article
Cite this article
Wagih, O., Reimand, J. & Bader, G. MIMP: predicting the impact of mutations on kinase-substrate phosphorylation. Nat Methods 12, 531–533 (2015). https://doi.org/10.1038/nmeth.3396
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3396
This article is cited by
-
KSP: an integrated method for predicting catalyzing kinases of phosphorylation sites in proteins
BMC Genomics (2020)
-
Using phosphoproteomics data to understand cellular signaling: a comprehensive guide to bioinformatics resources
Clinical Proteomics (2020)
-
Systematic analysis of the intersection of disease mutations with protein modifications
BMC Medical Genomics (2019)
-
Adipocyte-secreted BMP8b mediates adrenergic-induced remodeling of the neuro-vascular network in adipose tissue
Nature Communications (2018)
-
A Methodological Assessment and Characterization of Genetically-Driven Variation in Three Human Phosphoproteomes
Scientific Reports (2018)