Abstract
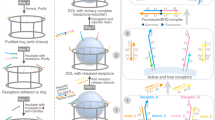
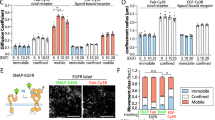
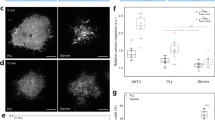
The spatial organization of membrane-bound ligands is thought to regulate receptor-mediated signaling. However, direct regulation of receptor function by nanoscale distribution of ligands has not yet been demonstrated, to our knowledge. We developed rationally designed DNA origami nanostructures modified with ligands at well-defined positions. Using these 'nanocalipers' to present ephrin ligands, we showed that the nanoscale spacing of ephrin-A5 directs the levels of EphA2 receptor activation in human breast cancer cells. Furthermore, we found that the nanoscale distribution of ephrin-A5 regulates the invasive properties of breast cancer cells. Our ligand nanocaliper approach has the potential to provide insight into the roles of ligand nanoscale spatial distribution in membrane receptor–mediated signaling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Casaletto, J.B. & McClatchey, A.I. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 12, 387–400 (2012).
Salaita, K. et al. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science 327, 1380–1385 (2010).
Lohmüller, T. et al. Supported membranes embedded with fixed arrays of gold nanoparticles. Nano Lett. 11, 4912–4918 (2011).
Holmberg, J. et al. Ephrin-A2 reverse signaling negatively regulates neural progenitor proliferation and neurogenesis. Genes Dev. 19, 462–471 (2005).
Pasquale, E.B. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180 (2010).
Miao, H. et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 16, 9–20 (2009).
Batlle, E. et al. EphB receptor activity suppresses colorectal cancer progression. Nature 435, 1126–1130 (2005).
Genander, M. et al. Dissociation of EphB2 signaling pathways mediating progenitor cell proliferation and tumor suppression. Cell 139, 679–692 (2009).
Bethani, I., Skånland, S.S., Dikic, I. & Acker-Palmer, A. Spatial organization of transmembrane receptor signalling. EMBO J. 29, 2677–2688 (2010).
Davis, S. et al. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 266, 816–819 (1994).
Wykosky, J. et al. Soluble monomeric EphrinA1 is released from tumor cells and is a functional ligand for the EphA2 receptor. Oncogene 27, 7260–7273 (2008).
Stein, E. et al. Eph receptors discriminate specific ligand oligomers to determine alternative signaling complexes, attachment, and assembly responses. Genes Dev. 12, 667–678 (1998).
Egea, J. et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron 47, 515–528 (2005).
Seeman, N.C. Nanomaterials based on DNA. Annu. Rev. Biochem. 79, 65–87 (2010).
Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Högberg, B. & Olin, H. DNA-scaffolded nanoparticle structures. J. Phys. Conf. Ser. 61, 458–462 (2007).
Voigt, N.V. et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 5, 200–203 (2010).
Selmi, D.N. et al. DNA-templated protein arrays for single-molecule imaging. Nano Lett. 11, 657–660 (2011).
Rinker, S., Ke, Y., Liu, Y., Chhabra, R. & Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol. 3, 418–422 (2008).
Park, S.H. et al. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 5, 729–733 (2005).
Derr, N.D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012).
Douglas, S.M., Bachelet, I. & Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Douglas, S.M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Andersen, E.S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
Abulrob, A. et al. Nanoscale imaging of epidermal growth factor receptor clustering: effects of inhibitors. J. Biol. Chem. 285, 3145–3156 (2010).
Lajoie, P. et al. Plasma membrane domain organization regulates EGFR signaling in tumor cells. J. Cell Biol. 179, 341–356 (2007).
Castro, C.E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Söderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Pitulescu, M.E. & Adams, R.H. Eph/ephrin molecules—a hub for signaling and endocytosis. Genes Dev. 24, 2480–2492 (2010).
Zhuang, G., Hunter, S., Hwang, Y. & Chen, J. Regulation of EphA2 receptor endocytosis by SHIP2 lipid phosphatase via phosphatidylinositol 3-Kinase-dependent Rac1 activation. J. Biol. Chem. 282, 2683–2694 (2007).
Hiramoto-Yamaki, N. et al. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J. Cell Biol. 190, 461–477 (2010).
Macrae, M. et al. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell 8, 111–118 (2005).
Himanen, J.P. et al. Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. USA 107, 10860–10865 (2010).
Seiradake, E., Harlos, K., Sutton, G., Aricescu, A.R. & Jones, E.Y. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat. Struct. Mol. Biol. 17, 398–402 (2010).
Wimmer-Kleikamp, S.H., Janes, P.W., Squire, A., Bastiaens, P.I.H. & Lackmann, M. Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J. Cell Biol. 164, 661–666 (2004).
Mitov, M.I., Greaser, M.L. & Campbell, K.S. GelBandFitter—a computer program for analysis of closely spaced electrophoretic and immunoblotted bands. Electrophoresis 30, 848–851 (2009).
Coffman, K.T. et al. Differential EphA2 epitope display on normal versus malignant cells. Cancer Res. 63, 7907–7912 (2003).
Acknowledgements
We thank U. Lendahl and his lab (Karolinska Institutet) for reagents and discussions, J. Avila-Cariño for help with the FACS experiments, O. Shupliakov for help with TEM, G. Bernardinelli for help with rendering ephrin-A5 and Y.-X. Zhao for help with initial experiments. This work was funded through grants from the Swedish Research Council to B.H. (repatriation grant 2010-6296 and project grant 2010-5060) and by the Strategic Research Program in Stem Cell Research and Regenerative Medicine at Karolinska Institutet (StratRegen), Sweden (A.I.T.). B.H. received startup funding from Carl Bennet AB, Karolinska Institutet and the Swedish Governmental Agency for Innovation Systems (Vinnova). V.L. and A.S. were supported by KID doctoral fellowships from the Karolinska Institutet, Sweden.
Author information
Authors and Affiliations
Contributions
B.H. and A.I.T. conceived of and designed the study. V.L., A.S. and E.P. performed most of the experimental work. F.F. and E.B. contributed to nanostructure and conjugate production and nanocaliper characterization. A.A.-A., A.H. and A.B. contributed to cell culture and the setting up and validating of the PLA assay. A.B. performed immunoprecipitation and immunoblotting experiments. V.L., A.S., B.H. and A.I.T. wrote the manuscript. All authors contributed to manuscript proofing and discussion.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16 and Supplementary Table 1 (PDF 9518 kb)
Supplementary Data
Raw FACS source data for Figure 2b. (ZIP 738 kb)
Rights and permissions
About this article
Cite this article
Shaw, A., Lundin, V., Petrova, E. et al. Spatial control of membrane receptor function using ligand nanocalipers. Nat Methods 11, 841–846 (2014). https://doi.org/10.1038/nmeth.3025
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3025
This article is cited by
-
Soluble and multivalent Jag1 DNA origami nanopatterns activate Notch without pulling force
Nature Communications (2024)
-
DNA-functionalized artificial mechanoreceptor for de novo force-responsive signaling
Nature Chemical Biology (2024)
-
DNA T-shaped crossover tiles for 2D tessellation and nanoring reconfiguration
Nature Communications (2023)
-
Functionalizing DNA origami to investigate and interact with biological systems
Nature Reviews Materials (2022)
-
Designer DNA nanostructures for viral inhibition
Nature Protocols (2022)