Abstract
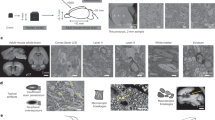
The development of methods for imaging large contiguous volumes with the electron microscope could allow the complete mapping of a whole mouse brain at the single-axon level. We developed a method based on prolonged immersion that enables staining and embedding of the entire mouse brain with uniform myelin staining and a moderate preservation of the tissue's ultrastructure. We tested the ability to follow myelinated axons using serial block-face electron microscopy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kasthuri, N. & Lichtman, J.W. Nat. Methods 4, 307–308 (2007).
Bohland, J.W. et al. PLoS Comput. Biol. 5, e1000334 (2009).
Tsai, P.S. et al. J. Neurosci. 29, 14553–14570 (2009).
Sabatini, D.D., Bensch, K. & Barrnett, R.J. J. Cell Biol. 17, 19–58 (1963).
Dalton, A.J., Kahler, H., Striebich, M.J. & Lloyd, B. J. Natl. Cancer Inst. 11, 439–461 (1950).
Palay, S.L., McGee-Russell, S.M., Gordon, S. Jr. & Grillo, M.A. J. Cell Biol. 12, 385–410 (1962).
Seligman, A.M., Wasserkrug, H.L. & Hanker, J.S. J. Cell Biol. 30, 424–432 (1966).
Li, A. et al. Science 330, 1404–1408 (2010).
Chung, J.R. et al. Front. Neuroinform. 5, 29 (2011).
Dempster, W.T. Am. J. Anat. 107, 59–72 (1960).
Hagström, L. & Bahr, G.F. Histochem. Cell Biol. 2, 1–4 (1960).
Bozzola, J.J. & Russell, L.D. Electron Microscopy: Principles and Techniques for Biologists. (Jones & Bartlett Learning, 1999).
Hayat, M.A. Principles and Techniques of Electron Microscopy: Biological Applications, 4th edn. (Cambridge University Press, 2000).
Van Holde, K.E. J. Phys. Chem. 63, 1574–1577 (1959).
Bahr, G.F. Exp. Cell Res. 7, 457–479 (1954).
Guha, P.C. & De, S.C. J. Chem. Soc. Trans. 125, 1215–1218 (1924).
De Bruijn, W.C. in Proc. 4th Eur. Reg. Conf. Electron Microsc. (ed. Bocciarelli, D.S.) 11–65 (Rome: Tipografia Polyglotta Vaticana, 1968).
Karnovsky, M.J. in Proc. 11th Meet. Am. Soc. Cell Biol. 146 (1971).
Seligman, A.M., Hanker, J.S., Wasserkrug, H., Dmochowski, H. & Katzoff, L. J. Histochem. Cytochem. 13, 629–639 (1965).
Quarles, R.H., Macklin, W.B. & Morell, P. in Basic Neurochemistry: Molecular, Cellular, and Medical Aspects, 7th edn. (eds. Siegel, G., Albers, R.W., Brady, S. & Price, D.) Ch. 4, 51–72 (Academic, 2006).
Spurr, A.R. J. Ultrastruct. Res. 26, 31–43 (1969).
Kushida, H. J. Electron Microsc. (Tokyo) 23, 197 (1974).
Partadiredja, G., Miller, R. & Oorschot, D.E. J. Neurocytol. 32, 1165–1179 (2003).
Sturrock, R.R. Neuropathol. Appl. Neurobiol. 6, 415–420 (1980).
Denk, W. & Horstmann, H. PLoS Biol. 2, e329 (2004).
Terzakis, J.A. J. Ultrastruct. Res. 22, 168–184 (1968).
Walton, J. J. Histochem. Cytochem. 27, 1337–1342 (1979).
Willingham, M.C. & Rutherford, A.V. J. Histochem. Cytochem. 32, 455–460 (1984).
Deerinck, T.J. et al. Microsc. Microanal. 16 (suppl. 2), 1138–1139 (2010).
Luft, J.H. Anat. Rec. 133, 305 (1959).
Binding, J., Mikula, S. & Denk, W. Microsc. Microanal. (in the press).
Holland, P.W. & Welsch, R.E. Commun. Stat. Theory Methods 6, 813–827 (1977).
Baumann, M. et al. in Proc. 6th PIMS Ind. Problem Solving Workshop (ed. Macki, J.) Ch. 1, 1–25 (PIMS, 2002).
Mikula, S., Trotts, I., Stone, J.M. & Jones, E.G. Neuroimage 35, 9–15 (2007).
Reynolds, E.S. J. Cell Biol. 17, 208–212 (1963).
Franklin, K.B.J. & Paxinos, G. The Mouse Brain in Stereotaxic Coordinates 3rd edn. (Academic, 2008).
Acknowledgements
We thank I. Sonntag and M. Helmstaedter for help with traceability analysis, S. Hillmer and U. Mersdorf for help with transmission electron microscopy, K. Briggman, S.K. Mikula and A. Scherbarth for help with staining procedures, B. Titze for help with conductive coating, J. Tritthardt for developing electronic circuits and M. Mueller for help with scanning electron microscopy–related software. We also thank the following student tracers: M. Diemer, C. Domnick, J. Hanne, P. Hofmann, A. Ivanova, H. Jakobi, A. Klein, J. Löffler, J. Nassel, J. Trendel and P. Weber. This work was supported by the Max Planck Society and the Deutsche Forschungsgemeinschaft (DFG).
Author information
Authors and Affiliations
Contributions
S.M. and W.D. designed the study and devised the analysis; S.M. carried out the experiments; J.B. and W.D. devised the aberration correction algorithm; S.M. analyzed the data; and S.M. and W.D. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
W.D. receives license income for 3View serial block-face electron microscopy technology from Gatan.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Tables 1–3 and Supplementary Protocol (PDF 663 kb)
Fly-through of a subregion from the internal capsule region of interest.
Voxel size is 40 nm isotropic (MOV 24047 kb)
Rotation animation of axon tracings from the ventroposterolateral nucleus of the dorsal thalamus.
Nodes of Ranvier are indicated by small gray spheres (MOV 17598 kb)
Rights and permissions
About this article
Cite this article
Mikula, S., Binding, J. & Denk, W. Staining and embedding the whole mouse brain for electron microscopy. Nat Methods 9, 1198–1201 (2012). https://doi.org/10.1038/nmeth.2213
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2213
This article is cited by
-
Volume electron microscopy
Nature Reviews Methods Primers (2022)
-
DeepACSON automated segmentation of white matter in 3D electron microscopy
Communications Biology (2021)
-
Protocol for preparation of heterogeneous biological samples for 3D electron microscopy: a case study for insects
Scientific Reports (2021)
-
Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues
Nature Communications (2020)
-
A carbon nanotube tape for serial-section electron microscopy of brain ultrastructure
Nature Communications (2018)