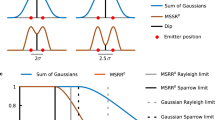
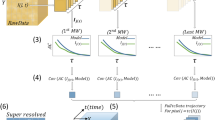
Abstract
In super-resolution microscopy methods based on single-molecule switching, the rate of accumulating single-molecule activation events often limits the time resolution. Here we developed a sparse-signal recovery technique using compressed sensing to analyze images with highly overlapping fluorescent spots. This method allows an activated fluorophore density an order of magnitude higher than what conventional single-molecule fitting methods can handle. Using this method, we demonstrated imaging microtubule dynamics in living cells with a time resolution of 3 s.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Huang, B., Babcock, H. & Zhuang, X. Cell 143, 1047–1058 (2010).
Hell, S.W. Nat. Methods 6, 24–32 (2009).
Rust, M.J., Bates, M. & Zhuang, X. Nat. Methods 3, 793–795 (2006).
Betzig, E. et al. Science 313, 1642–1645 (2006).
Hess, S.T., Girirajan, T.P.K. & Mason, M.D. Biophys. J. 91, 4258–4272 (2006).
Shroff, H., Galbraith, C.G., Galbraith, J.A. & Betzig, E. Nat. Methods 5, 417–423 (2008).
Jones, S.A., Shim, S.H., He, J. & Zhuang, X. Nat. Methods 8, 499–505 (2011).
Holden, S.J., Uphoff, S. & Kapanidis, A.N. Nat. Methods 8, 279–280 (2011).
Huang, F., Schwartz, S.L., Byars, J.M. & Lidke, K.A. Biomed. Opt. Express 2, 1377–1393 (2011).
Quan, T. et al. Opt. Express 19, 16963–16974 (2011).
Cox, S. et al. Nat. Methods 9, 195–200 (2012).
Candès, E.J., Romberg, J. & Tao, T. IEEE Trans. Inf. Theory 52, 489–509 (2004).
Zhu, L. et al. Phys. Med. Biol. 53, 6653–6672 (2008).
Bates, M., Huang, B., Dempsey, G.T. & Zhuang, X. Science 317, 1749–1753 (2007).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Science 319, 810–813 (2008).
Candès, E.J., Romberg, J. & Tao, T. Commun. Pure Appl. Math. 59, 1207–1223 (2005).
Zhu, L. & Xing, L. Med. Phys. 36, 1895–1905 (2009).
Mortensen, K.I., Churchman, L.S., Spudich, J.A. & Flyvbjerg, H. Nat. Methods 7, 377–381 (2010).
Grant, M. & Boyd, S. CVX: Matlab software for disciplined convex programming, version 1.21 <http://cvxr.com/cvx> (2011).
Huang, B., Jones, S.A., Brandenburg, B. & Zhuang, X. Nat. Methods 5, 1047–1052 (2008).
Acknowledgements
We thank E. Griffis and R. Vale (University of California, San Francisco) for generously providing the mEos2-tubulin S2 cells, and Q. Fan (Georgia Institute of Technology) for running the DAOSTORM code. L.Z. receives support from US National Institutes of Health 1R21EB012700-01 A1. B.H. receives support from the UCSF Program for Breakthrough Biomedical Research, Searle Scholarship, and Packard Fellowship for Science and Engineering.
Author information
Authors and Affiliations
Contributions
L.Z. and B.H. conceived the project and developed the algorithms, W.Z. performed the experiments, D.E. and B.H. analyzed the data, and L.Z. and B.H. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Note (PDF 725 kb)
Supplementary Video 1
STORM 'movie' of microtubules in a living Drosophila S2 cell stably expressing mEos2-fused tubulin, with a time resolution of 3 seconds. The movie is reconstructed from 4,349 camera frames (77 seconds) and plays 11 times as fast as real time. Three snapshots from the movie are shown in Figure 2b. Scale bar, 1 μm. (MOV 1944 kb)
Supplementary Software
Matlab code for STORM data analysis using compressed sensing. (ZIP 221 kb)
Rights and permissions
About this article
Cite this article
Zhu, L., Zhang, W., Elnatan, D. et al. Faster STORM using compressed sensing. Nat Methods 9, 721–723 (2012). https://doi.org/10.1038/nmeth.1978
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1978
This article is cited by
-
On the Determination of Lagrange Multipliers for a Weighted LASSO Problem Using Geometric and Convex Analysis Techniques
Applied Mathematics & Optimization (2024)
-
RegiSTORM: channel registration for multi-color stochastic optical reconstruction microscopy
BMC Bioinformatics (2023)
-
Super-resolution SRS microscopy with A-PoD
Nature Methods (2023)
-
Temporally resolved SMLM (with large PAR shift) enabled visualization of dynamic HA cluster formation and migration in a live cell
Scientific Reports (2023)
-
Single-frame deep-learning super-resolution microscopy for intracellular dynamics imaging
Nature Communications (2023)