Abstract
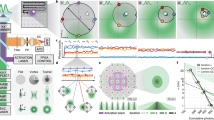
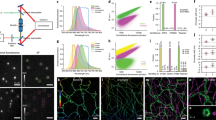
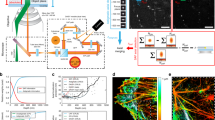
We demonstrate three-dimensional (3D) super-resolution microscopy in whole fixed cells using photoactivated localization microscopy (PALM). The use of the bright, genetically expressed fluorescent marker photoactivatable monomeric (m)Cherry (PA-mCherry1) in combination with near diffraction-limited confinement of photoactivation using two-photon illumination and 3D localization methods allowed us to investigate a variety of cellular structures at <50 nm lateral and <100 nm axial resolution. Compared to existing methods, we have substantially reduced excitation and bleaching of unlocalized markers, which allows us to use 3D PALM imaging with high localization density in thick structures. Our 3D localization algorithms, which are based on cross-correlation, do not rely on idealized noise models or specific optical configurations. This allows instrument design to be flexible. By generating appropriate fusion constructs and expressing them in Cos7 cells, we could image invaginations of the nuclear membrane, vimentin fibrils, the mitochondrial network and the endoplasmic reticulum at depths of greater than 8 μm.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
04 March 2011
In the version of this article initially published online, the affiliation for Alipasha Vaziri was incorrect. The error has been corrected for the print, PDF and HTML versions of this article.
References
Larabell, C.A. & Le Gros, M.A. X-ray tomography generates 3-D reconstructions of the yeast, Saccharomyces cerevisiae, at 60-nm resolution. Mol. Biol. Cell 15, 957–962 (2004).
Hohmann-Marriott, M.F. et al. Nanoscale 3D cellular imaging by axial scanning transmission electron tomography. Nat. Methods 6, 729–731 (2009).
Gustafsson, M.G.L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Gustafsson, M.G.L. et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 (2008).
Donnert, G. et al. Macromolecular-scale resolution in biological fluorescence microscopy. Proc. Natl. Acad. Sci. USA 103, 11440–11445 (2006).
Schmidt, R. et al. Spherical nanosized focal spot unravels the interior of cells. Nat. Methods 5, 539–544 (2008).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S.T., Girirajan, T.P.K. & Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Shtengel, G. et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl. Acad. Sci. USA 106, 3125–3130 (2009).
Axelrod, D. Total internal reflection fluorescence microscopy in cell biology. Traffic 2, 764–774 (2001).
Vaziri, A., Tang, J., Shroff, H. & Shank, C.V. Multilayer three-dimensional super resolution imaging of thick biological samples. Proc. Natl. Acad. Sci. USA 105, 20221–20226 (2008).
Fölling, J. et al. Photochromic rhodamines provide nanoscopy with optical sectioning. Angew. Chem. Int. Edn. Engl. 46, 6266–6270 (2007).
Mlodzianoski, M.J., Juette, M.F., Beane, G.L. & Bewersdorf, J. Experimental characterization of 3D localization techniques for particle-tracking and super-resolution microscopy. Opt. Express 17, 8264–8277 (2009).
Huang, B., Jones, S.A., Brandenburg, B. & Zhuang, X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods 5, 1047–1052 (2008).
Subach, F.V. et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009).
Denk, W., Strickler, J.H. & Webb, W.W. Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990).
Zhu, G., van Howe, J., Durst, M., Zipfel, W. & Xu, C. Simultaneous spatial and temporal focusing of femtosecond pulses. Opt. Express 13, 2153–2159 (2005).
Oron, D., Tal, E. & Silberberg, Y. Scanningless depth-resolved microscopy. Opt. Express 13, 1468–1476 (2005).
Ando, R., Mizuno, H. & Miyawaki, A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306, 1370–1373 (2004).
Tal, E., Oron, D. & Silberberg, Y. Improved depth resolution in video-rate line-scanning multiphoton microscopy using temporal focusing. Opt. Lett. 30, 1686–1688 (2005).
Shroff, H. et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc. Natl. Acad. Sci. USA 104, 20308–20313 (2007).
Juette, M.F. et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5, 527–529 (2008).
Pavani, S.R.P. et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA 106, 2995–2999 (2009).
Smith, C.S., Joseph, N., Rieger, B. & Lidke, K.A. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods 7, 373–375 (2010).
Pertsinidis, A., Zhang, Y. & Chu, S. Subnanometre single-molecule localization, registration, and distance measurements. Nature 466, 647–651 (2010).
Arimoto, R. & Murray, J.M. A common aberration with water-immersion objective lenses. J. Microsc. 216, 49–51 (2004).
Guizar-Sicairos, M., Thurman, S.T. & Fienup, J.R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
Oliphant, T. Python for scientific computing. Comput. Sci. Eng. 9, 10–20 (2007).
Gupton, S.L., Collings, D.A. & Allen, N.S. Endoplasmic reticulum targeted GFP reveals ER organization in tobacco NT-1 cells during cell division. Plant Physiol. Biochem. 44, 95–105 (2006).
Yoon, M., Moir, R.D., Prahlad, V. & Goldman, R.D. Motile properties of vimentin intermediate filament networks in living cells. J. Cell Biol. 143, 147–157 (1998).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E.H.K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Ritter, J.G., Veith, R., Jan-Peter, S. & Kubitscheck, U. High-contrast single-particle tracking by selective focal plane illumination microscopy. Opt. Express 16, 7142–7152 (2008).
Mortensen, K.I., Churchman, L.S., Spudich, J.A. & Flyvbjerg, H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods 7, 377–381 (2010).
Hunter, J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Acknowledgements
We thank N. Morgan and A. Gillespie for training and use of their spin coater; G. Patterson (National Institute of Biomedical Imaging and Bioengineering) for the gift of purified PA-mCherry1 and mEos2 and for the use of his cell culture facilities; A. Jin for measuring the thickness of our quantum dot films; E. Ramko for help with preparing the PA-mCherry1 fusion vectors; K. Kilborn (Intelligent Imaging Innovations) for loaning us the Vector Point Scanning 2P system; and S. Parekh and H. Eden for feedback and suggestions on the manuscript. This work was supported by the Intramural Research Programs of the National Institute of Biomedical Imaging and Bioengineering.
Author information
Authors and Affiliations
Contributions
A.G.Y., A.G. and H.S. conceived, designed and built the experimental setup. A.G.Y. wrote the analysis code. A.G. and H.S. collected the data. A.G.Y., A.G. and H.S. analyzed the data. M.W.D. and A.V. contributed reagents and materials. A.G.Y., M.W.D. and H.S. wrote the paper. All authors edited and refined the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16, Supplementary Table 1 and Supplementary Notes 1–3 (PDF 2999 kb)
Supplementary Video 1
z-stack of PA-mCherry1-mito fusions, to accompany Figure 3. Histogram bin size is 60 nm, individual frames are separated by 60 nm z steps. Smoothing of σ = 0.4 pixels in each dimension was applied before plotting data. (MOV 1173 kb)
Supplementary Video 2
z-stack of PA-mCherry1-ER fusions, to accompany Figure 4. Histogram bin size is 60 nm, individual frames are separated by 60 nm z steps. Smoothing of σ = 0.6 pixels in each dimension was applied before plotting data. (MOV 1519 kb)
Supplementary Video 3
z-stack of PA-mCherry1-vimentin fusions, to accompany Figure 5. Histogram bin size is 60 nm, individual frames are separated by 60 nm z steps. Smoothing of σ = 0.6 pixels in each dimension was applied before plotting data. (MOV 4243 kb)
Supplementary Video 4
z-stack of PA-mCherry1-lamin fusions, to accompany Figure 6. Histogram bin size is 50 nm, individual frames are separated by 50 nm z steps. Smoothing of σ = 0.75 pixels in each dimension was applied before plotting data. (MOV 7782 kb)
Supplementary Video 5
z-stack of PA-mCherry1-lamin fusions, extending over > 8.5 μm imaging depth. Histogram bin size is 60 nm, individual frames are separated by 60 nm z steps. Smoothing of σ = 0.75 pixels in each dimension was applied before plotting data. (MOV 3267 kb)
Supplementary Software
Supplementary Software (ZIP 51 kb)
Rights and permissions
About this article
Cite this article
York, A., Ghitani, A., Vaziri, A. et al. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods 8, 327–333 (2011). https://doi.org/10.1038/nmeth.1571
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1571
This article is cited by
-
High-speed super-resolution imaging of rotationally symmetric structures using SPEED microscopy and 2D-to-3D transformation
Nature Protocols (2021)
-
Potential quality improvement of stochastic optical localization nanoscopy images obtained by frame by frame localization algorithms
Scientific Reports (2020)
-
Super-resolution microscopy as a powerful tool to study complex synthetic materials
Nature Reviews Chemistry (2019)
-
Visualisation and analysis of hepatitis C virus non-structural proteins using super-resolution microscopy
Scientific Reports (2018)
-
Real-time 3D single-molecule localization using experimental point spread functions
Nature Methods (2018)