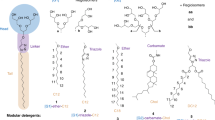
Abstract
The understanding of integral membrane protein (IMP) structure and function is hampered by the difficulty of handling these proteins. Aqueous solubilization, necessary for many types of biophysical analysis, generally requires a detergent to shield the large lipophilic surfaces of native IMPs. Many proteins remain difficult to study owing to a lack of suitable detergents. We introduce a class of amphiphiles, each built around a central quaternary carbon atom derived from neopentyl glycol, with hydrophilic groups derived from maltose. Representatives of this maltose–neopentyl glycol (MNG) amphiphile family show favorable behavior relative to conventional detergents, as manifested in multiple membrane protein systems, leading to enhanced structural stability and successful crystallization. MNG amphiphiles are promising tools for membrane protein science because of the ease with which they may be prepared and the facility with which their structures may be varied.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
09 November 2010
In the version of this article initially published online, Figure 1 contained errors (an incorrect number of carbons were drawn in the molecules). The error has been corrected for the print, PDF and HTML versions of this article.
References
Liu, J. & Rost, B. Comparing function and structure between entire proteomes. Protein Sci. 10, 1970–1979 (2001).
Lacapère, J.J., Pebay-Peyroula, E., Neumann, J.M. & Etchebest, C. Determining membrane protein structures: still a challenge! Trends Biochem. Sci. 32, 259–270 (2007).
Privé, G.G. Detergents for the stabilization and crystallization of membrane proteins. Methods 41, 388–397 (2007).
Schafmeister, C.E., Meircke, L.J.W. & Stroud, R.M. Structure at 2.5 Å of a designed peptide that maintains solubility of membrane proteins. Science 262, 734–738 (1993).
McGregor, C.-L. et al. Lipopeptide detergents designed for the structural study of membrane protein. Nat. Biotechnol. 21, 171–176 (2003).
Zhao, X. et al. Designer short peptide surfactants stabilize G protein-coupled receptor bovine rhodopsin. Proc. Natl. Acad. Sci. USA 103, 17707–17712 (2006).
Tribet, C., Audebert, R. & Popot, J.-L. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. USA 93, 15047–15050 (1996).
Popot, J.-L. Amphipols, nanodiscs, and fluorinated surfactants: three non-conventional approaches to studying membrane proteins in aqueous solutions. Annu. Rev. Biochem. 79, 737–775 (2010).
Nath, A., Atkins, W.M. & Sligar, S.G. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry 46, 2059–2069 (2007).
Borch, J. & Hamann, T. The nanodisc: a novel tool for membrane protein studies. Biol. Chem. 390, 805–814 (2009).
Breyton, C. et al. Micellar and biochemical properties of (hemi)fluorinated surfactants are controlled by the size of the polar head. Biophys. J. 97, 1077–1086 (2009).
Zhang, Q. et al. Designing facial amphiphiles for the stabilization of integral membrane protein. Angew. Chem. Int. Edn. 119, 7153–7155 (2007).
Hoffmann, R.W. Flexible molecules with defined shape-conformational design. Angew. Chem. Int. Edn. Engl. 31, 1124–1134 (1992).
McQuade, D.T. et al. Rigid amphiphiles for membrane protein manipulation. Angew. Chem. Int. Edn. 39, 758–761 (2000).
Chae, P.S., Wander, M.J., Bowling, A.P., Laible, P.D. & Gellman, S.H. Glycotripod amphiphiles for solubilization and stabilization of a membrane protein superassembly: importance of branching in the hydrophilic portion. ChemBioChem 9, 1706–1709 (2008).
Rosenbaum, D.M. et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318, 1266–1273 (2007).
Bassilana, M., Pourcher, T. & Lablanc, G. Melibiose permease of Escherichia coli . J. Biol. Chem. 263, 9663–9667 (1988).
Alexandrov, A.I., Mileni, M., Chien, E.Y., Hanson, M.A. & Stevens, R.C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008).
Horsefield, R., Iwata, S. & Byrne, B. Complex II from a structural perspective. Curr. Protein Pept. Sci. 5, 107–118 (2004).
Puustinen, A., Finel, M., Haltia, T., Gennis, R.B. & Wikström, M. Properties of the two terminal oxidases of Escherichia coli . Biochemistry 30, 3936–3942 (1991).
Newstead, S., Kim, H., von Heijne, G., Iwata, S. & Drew, D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA 104, 13936–13941 (2007).
Deckert, G. et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus . Nature 392, 353–358 (1998).
Yamashita, A., Singh, S.K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 (2005).
Quick, M. & Javitch, J.A. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl. Acad. Sci. USA 104, 3603–3608 (2007).
Hu, X.C., Ritz, T., Damjanovic, A., Authenrieth, F. & Schulten, K. Photosynthetic apparatus of purple bacteria. Q. Rev. Biophys. 35, 1–62 (2002).
Youvan, D.C., Ismail, S. & Bylina, E.J. Chromosomal deletion and plasmid complementation of the photosynthetic reaction center and light-harvesting genes from Rhodopseudomonas capsulata . Gene 38, 19–30 (1985).
Stroebel, D., Choquet, Y., Popot, J.-L. & Picot, D. An atypical haem in the cytochrome b 6 f complex. Nature 426, 413–418 (2003).
Rosenbaum, D.M., Rasmussen, S.G.F. & Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 459, 356–363 (2009).
Cherezov, V. et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007).
Hanson, M.A. et al. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16, 897–905 (2008).
Sanders, C.R. & Sonnichsen, F. Solution NMR of membrane proteins: practice and challenges. Magn. Reson. Chem. 44, S24–S40 (2006).
Barrera, N.P., Di Bartolo, N., Booth, P.J. & Robinson, C.V. Micelles protect membrane complexes from solution to vacuum. Science 321, 243–246 (2008).
Li, L. et al. Simple host-guest chemistry to modulate the process of concentration and crystallization of membrane proteins by detergent capture in a microfluidic device. J. Am. Chem. Soc. 130, 14324–14328 (2008).
Pourcher, T., Leclercq, S., Brandolin, G. & Leblanc, G. Melibiose permease of Escherichia coli: large scale purification and evidence that H+, Na+, and Li+ sugar symport is catalyzed by a single polypeptide. Biochemistry 34, 4412–4420 (1995).
Drew, D. et al. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae . Nat. Protoc. 3, 784–798 (2008).
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Collaborative Computational Project. Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D50, 760–763 (1994).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Bricogne, G. et al. BUSTER, version 2.8.0. (Global Phasing Ltd., Cambridge, UK, 2009).
Pierre, Y., Breyton, C., Kramers, D. & Popot, J.-L. Purification and characterization of the cytochrome b 6 f complex from Chlamydomonas reinhardtii . J. Biol. Chem. 270, 29342–29349 (1995).
Goren, M.A. & Fox, B.G. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr. Purif. 62, 171–178 (2008).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grant P01 GM75913 (S.H.G.), NS28471 (B.K.), by the Lundbeck Foundation (S.G.F.R., C.J.L. and U.G.), by the Danish National Research Council (C.J.L., U.G.), by the European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement no. HEALTH-F4-2007-201924, EDICT Consortium (K.G., U.G. and B.B.) and by NIH grant GM083118 and NIH Protein Structure Initiative grants U54 GM-074901 (J.L. Markley, PI; B.G.F.) and U54 GM094584 (B.G.F.). This work was also supported by grant no. R21HL087895 from the US National Heart, Lung, and Blood Institute, by the Texas Norman Hackerman Advanced Research Program under grant no. 010674-0034-2009 (to L.G.) and by the Center for Membrane Protein Research, Texas Tech University Health Sciences Center. R.R.R. was funded by the Defence Science and Technology Laboratory. We thank P. Laible (Argonne National Laboratory, Chicago) for supplying membrane preparations from R. capsulatus. We acknowledge SOLEIL (Saint-Aubin, France) for provision of synchrotron radiation facilities, and we would like to thank B. Guimaraes for assistance in using the beamline Proxima 1. We also thank R. Kaback (University of California, Los Angeles) and G. Leblanc (Institut de Biologie et Technologies–Saclay) for the MelB expression system. M.A.G. acknowledges support from the US National Science Foundation East Asia and Pacific Summer Institutes Fellowship program. We thank G. Cecchini (University of California, San Francisco) and J. Ruprecht (Medical Research Council Mitochondrial Biology Unit, Cambridge) for the purified SQR and the details of the SQR functional assay, and we acknowledge the assistance of P. Nixon in the analysis of the SQR functional data.
Author information
Authors and Affiliations
Contributions
P.S.C. designed the MNG amphiphiles, with contributions from S.G.F.R., B.K. and S.H.G. P.S.C. synthesized the amphiphiles. P.S.C., S.G.F.R., R.R.R., K.G., R.C., M.A.G., A.C.K., S.N., Y.P. and D.P. designed and performed the research and interpreted the data. C.J.L., D.D., B.G.F., L.G., U.G., J.-L.P., B.B., B.K. and S.H.G. contributed to experimental design and data interpretation. P.S.C. and S.H.G. wrote the manuscript, with oversight from S.G.F.R., R.R.R., K.G., R.C., M.A.G., A.C.K., S.N., C.J.L., Y.P., D.D., J.-L.P., D.P., B.G.F., L.G., U.G., B.B. and B.K.
Corresponding authors
Ethics declarations
Competing interests
P.S.C, S.G.F.R., B.K. and S.H.G. are co-inventors on a patent application that covers the MNG amphiphiles.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1 and 2, and Supplementary Note (PDF 864 kb)
Rights and permissions
About this article
Cite this article
Chae, P., Rasmussen, S., Rana, R. et al. Maltose–neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods 7, 1003–1008 (2010). https://doi.org/10.1038/nmeth.1526
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1526
This article is cited by
-
19F-NMR studies of the impact of different detergents and nanodiscs on the A2A adenosine receptor
Journal of Biomolecular NMR (2024)
-
Identity, structure, and function of the mitochondrial permeability transition pore: controversies, consensus, recent advances, and future directions
Cell Death & Differentiation (2023)
-
X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB
Communications Biology (2021)
-
G protein-coupled receptors: structure- and function-based drug discovery
Signal Transduction and Targeted Therapy (2021)
-
Biophysical and functional characterization of the human TAS1R2 sweet taste receptor overexpressed in a HEK293S inducible cell line
Scientific Reports (2021)