Abstract
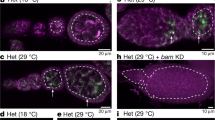
In Drosophila melanogaster, widely used mitotic recombination–based strategies generate mosaic flies with positive readout for only one daughter cell after division. To differentially label both daughter cells, we developed the twin spot generator (TSG) technique, which through mitotic recombination generates green and red twin spots that are detectable after the first cell division as single cells. We propose wide applications of TSG to lineage and genetic mosaic studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lee, T. & Luo, L. Neuron 22, 451–461 (1999).
Lee, T. & Luo, L. Trends Neurosci. 24, 251–254 (2001).
de la Cova, C., Abril, M., Bellosta, P., Gallant, P. & Johnston, L.A. Cell 117, 107–116 (2004).
Spradling, A.C. & Rubin, G.M. Science 218, 341–347 (1982).
Zong, H., Espinosa, J.S., Su, H.H., Muzumdar, M.D. & Luo, L. Cell 121, 479–492 (2005).
Golic, K.G. Science 252, 958–961 (1991).
Stern, C. Genetics 21, 625–730 (1936).
Cormack, B.P., Valdivia, R.H. & Falkow, S. Gene 173, 33–38 (1996).
Campbell, R.E. et al. Proc. Natl. Acad. Sci. USA 99, 7877–7882 (2002).
Harrison, D.A. & Perrimon, N. Curr. Biol. 3, 424–433 (1993).
Groth, A.C., Fish, M., Nusse, R. & Calos, M.P. Genetics 166, 1775–1782 (2004).
Bateman, J.R., Lee, A.M. & Wu, C.T. Genetics 173, 769–777 (2006).
McClure, K.D. & Schubiger, G. Development 132, 5033–5042 (2005).
Bateman, J.R. & Wu, C.T. Genetics 180, 1763–1766 (2008).
Rubin, G.M. & Spradling, A.C. Science 218, 348–353 (1982).
Spradling, A.C. & Rubin, G.M. Science 218, 341–347 (1982).
Bischof, J., Maeda, R.K., Hediger, M., Karch, F. & Basler, K. Proc. Natl. Acad. Sci. USA 104, 3312–3317 (2007).
Chou, T.B. & Perrimon, N. Genetics 144, 1673–1679 (1996).
Acknowledgements
We thank J. Zirin and M. Packard for confocal expertise, P. Bradley for molecular biology advice, F. Karch (University of Geneva) for providing flies carrying integrase and T.S. Griffin for contributing the technique acronym. This work was supported by grants from the US National Institutes of Health (1 RO1 GM61936, C.-t.W.; RO1 GM058282, G.S.; and RO1 GM084947, N.P.) and a Ruth L. Kirschstein National Research Service award (1 F32 GM67460, J.R.B.); the US National Science Foundation (A.M.H.); the Swiss National Science Foundation (PBBE33-121069, M.B.) and the Leukemia and Lymphoma Society (C.B.).
Author information
Authors and Affiliations
Contributions
R.G., design and execution of constructs, vectors and first-round proof-of-principle experiments, verification of TSG lines and writing of manuscipt. A.S., M.B. and G.S., clonal separation experiments and description in manuscript. R.B., design and generation of TSG transgenic fly lines and modification of target genomic lines. A.d.V.R., validation of TSG in fly brain. C.B., molecular biology expertise and tissue culture strategy. A.M.H., validation of TSG in imaginal discs. J.R.B., design and generation of target genomic lines. C.V., fly injections. E.H., tissue culture assays. D.G., acquisition and analysis of confocal images. C.D., G.S., C.-t.W. and N.P. co-directed the project. C.-t.W. suggested and funded the RMCE approach. N.P. provided US laboratory facilities and materials and conceived the TSG strategy.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–5 and Supplementary Note (PDF 3410 kb)
Rights and permissions
About this article
Cite this article
Griffin, R., Sustar, A., Bonvin, M. et al. The twin spot generator for differential Drosophila lineage analysis. Nat Methods 6, 600–602 (2009). https://doi.org/10.1038/nmeth.1349
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1349
This article is cited by
-
Investigating Adult Stem Cells Through Lineage analyses
Stem Cell Reviews and Reports (2022)
-
Deciphering neural heterogeneity through cell lineage tracing
Cellular and Molecular Life Sciences (2021)
-
Reply to ‘A refutation to ‘A new A-P compartment boundary and organizer in holometabolous insect wings’
Scientific Reports (2019)
-
Live imaging of stem cells in the germarium of the Drosophila ovary using a reusable gas-permeable imaging chamber
Nature Protocols (2018)
-
Alternative direct stem cell derivatives defined by stem cell location and graded Wnt signalling
Nature Cell Biology (2017)