Abstract
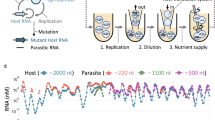
Self-replication and evolution under selective pressure are inherent phenomena in life, and but few artificial systems exhibit these phenomena1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17. We have designed a system of DNA origami rafts that exponentially replicates a seed pattern, doubling the copies in each diurnal-like cycle of temperature and ultraviolet illumination, producing more than 7 million copies in 24 cycles. We demonstrate environmental selection in growing populations by incorporating pH-sensitive binding in two subpopulations. In one species, pH-sensitive triplex DNA bonds enable parent–daughter templating, while in the second species, triplex binding inhibits the formation of duplex DNA templating. At pH 5.3, the replication rate of species I is ∼1.3–1.4 times faster than that of species II. At pH 7.8, the replication rates are reversed. When mixed together in the same vial, the progeny of species I replicate preferentially at pH 7.8; similarly at pH 5.3, the progeny of species II take over the system. This addressable selectivity should be adaptable to the selection and evolution of multi-component self-replicating materials in the nanoscopic-to-microscopic size range.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lincoln, T. A. & Joyce, G. F. Self-sustained replication of an RNA enzyme. Science 323, 1229–1232 (2009).
Wintner, E. A., Conn, M. M. & Rebek, J. Jr Studies in molecular replication. Acc. Chem. Res. 27, 198–203 (1994).
Mast, C. B., Schink, S., Gerland, U. & Braun, D. Escalation of polymerization in a thermal gradient. Proc. Natl Acad. Sci. USA 110, 8030–8035 (2013).
Schulman, R., Yurke, B. & Winfree, E. Robust self-replication of combinatorial information via crystal growth and scission. Proc. Natl Acad. Sci. USA 109, 6405–6410 (2012).
Lin, C. et al. In vivo cloning of artificial DNA nanostructures. Proc. Natl Acad. Sci. USA 105, 17626–17631 (2008).
Lee, D. H., Severin, K., Yokobayashi, Y. & Ghadiri, M. R. Emergence of symbiosis in peptide self-replication through a peptide hypercyclic network. Nature 390, 591–594 (1997).
Eckardt, L. H. et al. Chemical copying of connectivity. Nature 420, 286 (2002).
Wang, T. et al. Self-replication of information-bearing nanoscale patterns. Nature 478, 225–228 (2011).
Leunissen, M. E. et al. Towards self-replicating materials of DNA-functionalized colloids. Soft Matter 5, 2422–2430 (2009).
Levy, M. & Ellington, A. D. Exponential growth by cross-catalytic cleavage of deoxyribozymogens. Proc. Natl Acad. Sci. USA 100, 6416–6421 (2003).
Wochner, A., Attwater, J., Coulson, A. & Holliger, P. Ribozyme-catalyzed transcription of an active ribozyme. Science 332, 209–212 (2011).
Kim, J., Lee, J., Hamada, S., Murata, S. & Park, S. H. Self-replication of DNA rings. Nat. Nanotech. 10, 528–533 (2015).
Vaidya, N. et al. Spontaneous network formation among cooperative RNA replicators. Nature 491, 72–77 (2012).
Adamala, K. & Szostak, J. W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 342, 1098–1100 (2013).
Li, T. & Nicolaou, K. C. Chemical self-replication of palindromic duplex DNA. Nature 369, 218–221 (1994).
Luther, A., Brandsch, R. & von Kiedrowski, G. Surface-promoted replication and exponential amplification of DNA analogues. Nature 396, 245–248 (1998).
Colomb-Delsuc, M., Mattia, E., Sadownik, J. W. & Otto, S. Exponential self-replication enabled through a fibre elongation/breakage mechanism. Nat. Commun. 6, 7427 (2015).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind to specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Szostak, J. W., Bartel, D. P. & Luisi, P. L. Synthesizing life. Nature 409, 387–390 (2001).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Yoshimura, R. & Fujimoto, K. Ultrafast reversible photo-cross-linking reaction: toward in situ DNA manipulation. Org. Lett. 10, 3227–3230 (2008).
Rusling, D. A. et al. Functionalizing designer DNA crystals with a triple-helical veneer. Angew. Chem. Int. Ed. 53, 3979–3982 (2014).
Amodio, A. et al. Rational design of pH-controlled DNA strand displacement. J. Am. Chem. Soc. 136, 16469–16472 (2014).
Wu, N. & Willner, I. pH-stimulated reconfiguration and structural isomerization of origami dimer and trimer systems. Nano Lett. 16, 6650–6655 (2016).
Adleman, L. M. Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994).
Sacanna, S., Irvine, W. T. M., Chaikin, P. M. & Pine, D. J. Lock and key colloids. Nature 464, 575–578 (2010).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Wei, B., Wang, Z. & Mi, Y. Uniquimer: a de novo DNA sequence generation computer software for DNA self-assembly. J. Comput. Theoret. Nanosci. 4, 133–141 (2007).
Acknowledgements
We would like to acknowledge W. Liu, D. Niu and Y. Zhang for useful discussions. This research has been supported primarily by DOE DE-SC0007991 (PMC, NCS) for initiation, design, analysis and imaging, and partially by grant GBMF3849 from the Gordon and Betty Moore Foundation (PMC, NCS) for origami preparation and characterization, GM-29554 from NIGMS, grants CMMI-1120890 and CCF-1117210 from the NSF, MURI W911NF-11-1-0024 from ARO, grants N000141110729 and N000140911118 from ONR (NCS) for sequence design, DNA synthesis, purification and characterization. Y.M. and X.H. acknowledge the support by the Earmarked Grant from the University Grant Council of the Hong Kong Government, RGC 16302415, and the China 985 Grant of Tongji University. X.H. was supported partially by the MRSEC Program of the National Science Foundation under Award Number DMR-1420073 for pH-sensitive sequence design and characterization. We also wish to acknowledge the support of the National Science Foundation Academic Research Infrastructure program through Award No. CMMI-0957834.
Author information
Authors and Affiliations
Contributions
X.H., R.S., N.C.S. and P.M.C. designed the experiments. X.H., R.S. and R.Z. performed the experiments, and all of the authors analysed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1296 kb)
Rights and permissions
About this article
Cite this article
He, X., Sha, R., Zhuo, R. et al. Exponential growth and selection in self-replicating materials from DNA origami rafts. Nature Mater 16, 993–997 (2017). https://doi.org/10.1038/nmat4986
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat4986
This article is cited by
-
Proliferating coacervate droplets as the missing link between chemistry and biology in the origins of life
Nature Communications (2021)
-
DNA origami
Nature Reviews Methods Primers (2021)
-
Nanostructure evolution
Nature Materials (2017)