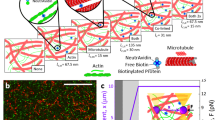
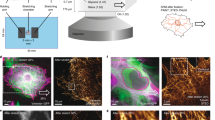
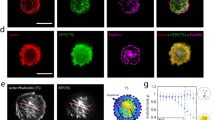
Abstract
Any macroscopic deformation of a filamentous bundle is necessarily accompanied by local sliding and/or stretching of the constituent filaments1,2. Yet the nature of the sliding friction between two aligned filaments interacting through multiple contacts remains largely unexplored. Here, by directly measuring the sliding forces between two bundled F-actin filaments, we show that these frictional forces are unexpectedly large, scale logarithmically with sliding velocity as in solid-like friction, and exhibit complex dependence on the filaments’ overlap length. We also show that a reduction of the frictional force by orders of magnitude, associated with a transition from solid-like friction to Stokes’s drag, can be induced by coating F-actin with polymeric brushes. Furthermore, we observe similar transitions in filamentous microtubules and bacterial flagella. Our findings demonstrate how altering a filament’s elasticity, structure and interactions can be used to engineer interfilament friction and thus tune the properties of fibrous composite materials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Vigolo, B. et al. Macroscopic fibres and ribbons of oriented carbon nanotubes. Science 290, 1331–1334 (2000).
Sanchez, T., Chen, D. T. N., DeCamp, S. J., Heymann, M. & Dogic, Z. Spontaneous motion in hierarchically assembled active matter. Nature 491, 431–434 (2012).
Kozlov, A. S., Baumgart, J., Risler, T., Versteegh, C. P. C. & Hudspeth, A. J. Forces between clustered stereocilia minimize friction in the ear on a subnanometre scale. Nature 474, 376–379 (2011).
Claessens, M., Bathe, M., Frey, E. & Bausch, A. R. Actin-binding proteins sensitively mediate F-actin bundle stiffness. Nature Mater. 5, 748–753 (2006).
Heussinger, C., Bathe, M. & Frey, E. Statistical mechanics of semiflexible bundles of wormlike polymer chains. Phys. Rev. Lett. 99, 048101 (2007)
Zimmerman, S. B. & Minton, A. P. Macromolecular crowding—biochemical, biophysical and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22, 27–65 (1993).
Rau, D. C., Lee, B. & Parsegian, V. A. Measurement of the repulsive force between poly-electrolyte molecules in ionic solution—hydration forces between parallel DNA double helices. Proc. Natl Acad. Sci. USA 81, 2621–2625 (1984).
Vanossi, A., Manini, N., Urbakh, M., Zapperi, S. & Tosatti, E. Colloquium: Modeling friction: From nanoscale to mesoscale. Rev. Mod. Phys. 85, 529–552 (2013).
Braun, O. M. & Kivshar, Y. S. Nonlinear dynamics of the Frenkel–Kontorova model. Phys. Rep. 306, 1–108 (1998).
Merkel, R., Nassoy, P., Leung, A., Ritchie, K. & Evans, E. Energy landscapes of receptor–ligand bonds explored with dynamic force spectroscopy. Nature 397, 50–53 (1999).
Gnecco, E. et al. Velocity dependence of atomic friction. Phys. Rev. Lett. 84, 1172–1175 (2000).
Muser, M. H., Urbakh, M. & Robbins, M. O. in Advances in Chemical Physics Vol. 126 (eds Prigogine, I. & Rice, S. A.) 187–272 (John Wiley, 2003).
Suda, H. Origin of friction derived from rupture dynamics. Langmuir 17, 6045–6047 (2001).
Holmes, K. C., Popp, D., Gebhard, W. & Kabsch, W. Atomic model of the actin filament. Nature 347, 44–49 (1990).
Kojima, H., Ishijima, A. & Yanagida, T. Direct measurement of stifness of single actin-filaments with and without topomyosin by in-vitro nanomanipulation. Proc. Natl Acad. Sci. USA 91, 12962–12966 (1994).
Lau, A. W. C., Prasad, A. & Dogic, Z. Condensation of isolated semi-flexible filaments driven by depletion interactions. Europhys. Lett. 87, 48006 (2009).
De Gennes, P. G. Maximum pull out force on DNA hybrids. C. R. Acad. Sci. IV 2, 1505–1508 (2001).
Hatch, K., Danilowicz, C., Coljee, V. & Prentiss, M. Demonstration that the shear force required to separate short double-stranded DNA does not increase significantly with sequence length for sequences longer than 25 base pairs. Phys. Rev. E 78, 011920 (2008).
Bormuth, V., Varga, V., Howard, J. & Schaffer, E. Protein friction limits diffusive and directed movements of kinesin motors on microtubules. Science 325, 870–873 (2009).
Choi, J. S. et al. Friction anisotropy-driven domain imaging on exfoliated monolayer graphene. Science 333, 607–610 (2011).
Forth, S., Hsia, K-C., Shimamoto, Y. & Kapoor, T. M. Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks. Cell 157, 420–432 (2014).
Li, G. L. & Tang, J. X. Diffusion of actin filaments within a thin layer between two walls. Phys. Rev. E 69, 061921 (2004).
Sanchez, T., Welch, D., Nicastro, D. & Dogic, Z. Cilia-like beating of active microtubule bundles. Science 333, 456–459 (2011).
Nogales, E., Wolf, S. G. & Downing, K. H. Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 (1998).
Klein, J., Kumacheva, E., Mahalu, D., Perahia, D. & Fetters, L. J. Reduction of frictional forces between solid-surfaces bearing polymer brushes. Nature 370, 634–636 (1994).
Akbulut, M., Belman, N., Golan, Y. & Israelachvili, J. Frictional properties of confined nanorods. Adv. Mater. 18, 2589–2592 (2006).
Mate, C. M., McClelland, G. M., Erlandsson, R. & Chiang, S. Atomic scale-friction of a tungsten tip on a graphite surface. Phys. Rev. Lett. 59, 1942–1945 (1987).
Luan, B. Q. & Robbins, M. O. The breakdown of continuum models for mechanical contacts. Nature 435, 929–932 (2005).
Yoshizawa, H., Chen, Y. L. & Israelachvili, J. Fundamental mechanism of interfacial friction. 1. Relation between adhesion and friction. J. Phys. Chem. 97, 4128–4140 (1993).
Van Alsten, J. & Granick, S. Molecular tribometry of ultrathin liquid-films. Phys. Rev. Lett. 61, 2570–2573 (1988).
Bohlein, T., Mikhael, J. & Bechinger, C. Observation of kinks and antikinks in colloidal monolayers driven across ordered surfaces. Nature Mater. 11, 126–130 (2012).
Acknowledgements
We acknowledge useful discussions with N. Upadhyaya, M. Hagan and R. Bruinsma. A.W., D.W. and W.S. were supported by National Science Foundation grants CMMI-1068566, NSF-MRI-0923057 and NSF-MRSEC-1206146. F.H. and Z.D. were supported by Department of Energy, Office of Basic Energy Sciences under Award DE-SC0010432TDD. V.V. acknowledges FOM and NWO for financial support. L.M. was supported by Harvard-NSF MRSEC and the MacArthur Foundation. We also acknowledge use of the Brandeis MRSEC optical microscopy facility (NSF-MRSEC-1206146).
Author information
Authors and Affiliations
Contributions
A.W. and Z.D. conceived the experiments. A.W. measured actin sliding friction and performed computer simulations. F.H., A.W. and D.W. performed microtubule sliding experiments. W.S. performed flagella sliding dynamics. A.W.C.L. developed a preliminary theoretical model that explains the velocity dependence of sliding friction. L.M. and V.V. developed the theoretical model that explains the dependence of sliding friction on overlap length. A.W., V.V., L.M. and Z.D. wrote the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1271 kb)
Supplementary Information
Supplementary Movie 1 (AVI 15866 kb)
Supplementary Information
Supplementary Movie 2 (AVI 3519 kb)
Supplementary Information
Supplementary Movie 3 (AVI 7260 kb)
Rights and permissions
About this article
Cite this article
Ward, A., Hilitski, F., Schwenger, W. et al. Solid friction between soft filaments. Nature Mater 14, 583–588 (2015). https://doi.org/10.1038/nmat4222
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat4222
This article is cited by
-
Interfacial Friction Evolution of DLC Films on the Fracturing Pump Plungers During the CO2 Fracturing Process: An Atomic Understanding from ReaxFF Simulations
Tribology Letters (2023)
-
Single-molecule force spectroscopy reveals the dynamic strength of the hair-cell tip-link connection
Nature Communications (2021)
-
Anillin propels myosin-independent constriction of actin rings
Nature Communications (2021)
-
Kinetics of motile solitons in nematic liquid crystals
Nature Communications (2020)
-
From mechanical resilience to active material properties in biopolymer networks
Nature Reviews Physics (2019)