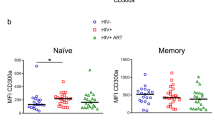
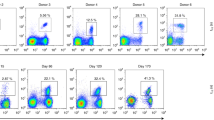
Abstract
The T-cell antigen receptor (TCR) repertoire was studied longitudinally by analyzing the varying lengths of the beta chain CDR3 hypervariable region during the course of HIV-1 infection and following combination antiretroviral therapy. Drastic restrictions in CD8+ T-cell repertoire usage were found at all stages of natural progression and persisted during the first six months of treatment. In contrast, significant CD4+ T-cell repertoire perturbations were not found in early stages of infection but correlated with progression to AIDS. Out of ten patients presenting with pretreatment perturbations, normalization of the CD4+ repertoire was observed in eight good responders, but not in two cases of unsuccessful therapy. These results indicate that, besides CD4+ cell count rise, an efficient control of HIV replication may allow qualitative modifications of the CD4+ repertoire balance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Lane, H.C. et al. Qualitative analysis of immune function in patients with the acquired immunodeficiency syndrome: Evidence for a selective defect in soluble antigen recognition. N. Engl. J. Med. 313, 79–84 (1985).
Roglic, M., Macphee, R.D., Duncan, S.R., Sattler, F.R. & Theophilopoulos, A.N. T cell receptor (TCR) BV gene repertoires and clonal expansions of CD4 cells in patients with HIV infections.Clin. Exp. Immunol. 107,21–30 (1997).
Fauci, A.S. Multifactorial nature of human immunodeficiency virus disease: Implications for therapy. Science 262, 1011–1018 (1993).
Perelson, A.S. et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387,188–191 (1997).
Connors, M. et al. HIV-1 infection induces changes in CD4+ T-cell phenotype and depletions within the CD4+ T-cell repertoire that are not immediately restored by antiviral or immune-based therapies. Nature Med. 3,533–540 (1997).
Davis, M.M. & Bjorkman, P.J. T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402(1988).
Rowen, L., Koop, B.F. & Hood, L. The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science 272,1755–1762 (1996).
Kepler, T.B., Borrero, M., Rugerio, B., McCray, S.K. & Clarke, S.H. Interdependence of N nucleotide addition and recombination site choice in V(D)J rearrangement. J. Immunol. 157,4451–4457(1996).
Jores, R. & Meo, T. Few V gene segments dominate the T cell receptor beta-chain repertoire of the human thymus. J. Immunol. 151, 6110–6122(1993).
Pannetier, C. et al. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc. Natl. Acad. Sci. USA 90, 4319–323 (1993).
Carmichael, A., Jin, X., Sissons, P. & Borysiewicz, L. Quantitative analysis of the human immunodeficiency virus type 1 (HIV-1 )-specific cytotoxic T lymphocyte (CTL) response at different stages of HIV-1 infection: Differential CTL responses to HIV-1 and Epstein-Barr virus in late disease. J. Exp. Med. 177, 249–256 (1993).
Bahadoran, P. et al. CD4+ T cells of human immunodeficiency virus-infected infants. Eur. J. Immunol. 23, 2041–2044 (1993).
Posnett, D.N., Kabak, S., Hodtsev, A.S., Goldberg, E.A. & Asch, A. T-cell antigen receptor V beta subsets are not preferentially deleted in AIDS. AIDS 7,625–631 (1993).
Pannetier, C., Even, J. & Kourilsky, P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol. Today 16, 176–181 (1995).
Jorgensen, J.L., Esser, U., de St. Groth, B.F., Reay, P.A., & Davis, M.M. Mapping T-cell re-ceptor-peptide contacts by variant peptide immunization of single-chain transgenics.. Nature 355, 224–230 (192).
Akolkar, P.N., Gulwani-Akolkar, B., Pergolizzi, R., Bigler, R.D. & Silver, J. Influence of HLA genes on T cell receptor V segment frequencies and expression levels in peripheral blood lymphocytes. J. Immunol. 150, 2761–2773 (1993).
Genevee, C. et al. An experimentally validated panel of subfamily-specific oligonu-cleotide primers (V alpha 1 -w29/V beta 1 -w24) for the study of human T cell receptor variable V gene segment usage by polymerase chain reaction. Eur. J. Immunol. 22 1261–1269 (1992).
Malhotra, U., Spielman, R. & Concannon, P. Variability in T cell receptor V beta gene usage in human peripheral blood lymphocytes. Studies of identical twins, siblings, and insulin-dependent diabetes mellitus patients. J. Immunol. 149 1802–1808 (1992).
Levraud, J.P., Pannetier, C., Langlade-Demoyen, P., Brichard, V. & Kourilsky, P. Recurrent T cell receptor rearrangements in the cytotoxic T lymphocyte response in vivo against the p815 murine tumor. J. Exp. Med. 183, 439–449 (1996).
Fitzgerald, J.E. et al. Analysis of clonal CD8+ T cell expansions in normal individuals and patients with rheumatoid arthritis. J. Immunol. 154, 3538–3547 (1995).
Mellors, J.W. et al. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272, 1167–1170 (1996).
Musette, P. et al. The J beta segment of the T cell receptor contributes to the V beta-specific T cell expansion caused by staphylococcal enterotoxin B and Urtica dioica super-antigens. Eur. J. Immunol. 26, 618–622 (1996).
Posnett, D.N., Sinha, R., Kabak, S. & Russo, C. Clonal populations of T cells in normal elderly humans: The T cell equivalent to “benign monoclonal gammapathy”. J. Exp. Med 179, 609–618 (1994).
Morley, J.K., Batliwalla, F.M., Hingorani, R. & Gregersen, P.K. Oligoclonal CD8+ T cells are preferentially expanded in the CD57+ subset. J. Immunol. 154, 6182–6190 (1995).
Schwab, R. et al. Expanded CD4+ and CD8+ T cell clones in elderly humans. J. Immunol. 158, 4493–499 (1997).
Hadida, F. et al. Carboxyl-terminal and central regions of human immunodeficiency virus-1 NEF recognized by cytotoxic T lymphocytes from lymphoid organs: An in vitro limiting dilution analysis. J. Clin. Invest. 89, 53–60 (1992).
Haas, G. et al. Dynamics of viral variants in HIV-1 Nef and specific cytotoxic T lymphocytes in vivo. J. Immunol. 157, 4212–4221 (1996).
Kalams, S.A. et al. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J. Exp. Med 179, 1261–1271 (1994).
Pantaleo, G. et al. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature 370, 463–467 (1994).
Tanchot, C., Lemonnier, F.A., Pérarneau, B., Freitas, A.A., Rocha, B. Differential recruitment for survival and proliferation of CD8 naive and memory T cells. Science 276, 2057–2062 (1997).
Sprent, J., Tough, D.F. & Sun, S. Factors controlling the turnover of T memory cells. Immunol. Rev. 156, 79–85 (1997).
Neumann, A.U.N. & Gorochov, G. HIV antivirals and immune recovery [letter].Nature Med. 3, 703–704 (1997).
Autran, B. et al. Positive effects of combined anti-retroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science 277, 112–116 (1997).
Ho, D.D. et al. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126 (1995).
Gorochov, G. et al. Oligoclonal expansion of CD8+ CD57+ T cells with restricted T-cell receptor beta chain variability after bone marrow transplantation. Blood 83, 587–595 (1994).
Kabat, E.A., Wu, T.T., Perry, H.M., Gottesman, K.S. & Foeller, C. Sequences of proteins of immunological interest. (National Institute of Health, Bethesda, 1991).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gorochov, G., Neumann, A., Kereveur, A. et al. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat Med 4, 215–221 (1998). https://doi.org/10.1038/nm0298-215
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nm0298-215
This article is cited by
-
Lymphocyte homeostasis is maintained in perinatally HIV-infected patients after three decades of life
Immunity & Ageing (2019)
-
Enhanced expression of PD-1 and other activation markers by CD4+ T cells of young but not old patients with metastatic melanoma
Cancer Immunology, Immunotherapy (2018)
-
T follicular helper and T follicular regulatory cells have different TCR specificity
Nature Communications (2017)
-
B- and T-lymphocyte number and function in HIV+/HIV− lymphoma patients treated with high-dose chemotherapy and autologous bone marrow transplantation
Scientific Reports (2016)
-
Serious Non-AIDS Events: Therapeutic Targets of Immune Activation and Chronic Inflammation in HIV Infection
Drugs (2016)