Abstract
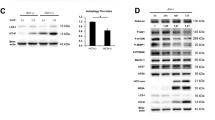
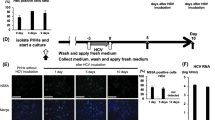
Hepatitis C virus (HCV) infects 200 million people globally, and 60–80% of cases persist as a chronic infection that will progress to cirrhosis and liver cancer in 2–10% of patients1,2,3. We recently demonstrated that HCV induces aberrant expression of two host microRNAs (miRNAs), miR-208b and miR-499a-5p, encoded by myosin genes in infected hepatocytes4. These miRNAs, along with AU-rich-element-mediated decay, suppress IFNL2 and IFNL3, members of the type III interferon (IFN) gene family, to support viral persistence. In this study, we show that miR-208b and miR-499a-5p also dampen type I IFN signaling in HCV-infected hepatocytes by directly down-regulating expression of the type I IFN receptor chain, IFNAR1. Inhibition of these miRNAs by using miRNA inhibitors during HCV infection increased expression of IFNAR1. Additionally, inhibition rescued the antiviral response to exogenous type I IFN, as measured by a marked increase in IFN-stimulated genes and a decrease in HCV load. Treatment of HCV-infected hepatocytes with type I IFN increased expression of myosins over HCV infection alone. Since these miRNAs can suppress type III IFN family members, these data collectively define a novel cross-regulation between type I and III IFNs during HCV infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142, 1264–1273.e1 (2012).
Lavanchy, D. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 17, 107–115 (2011).
Shepard, C.W., Finelli, L. & Alter, M.J. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5, 558–567 (2005).
McFarland, A.P. et al. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat. Immunol. 15, 72–79 (2014).
Liang, T.J. & Ghany, M.G. Current and future therapies for hepatitis C virus infection. N. Engl. J. Med. 368, 1907–1917 (2013).
Manns, M.P. & von Hahn, T. Novel therapies for hepatitis C—one pill fits all? Nat. Rev. Drug Discov. 12, 595–610 (2013).
Ghany, M.G., Nelson, D.R., Strader, D.B., Thomas, D.L. & Seeff, L.B. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology 54, 1433–1444 (2011).
Lawitz, E. et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N. Engl. J. Med. 368, 1878–1887 (2013).
Afdhal, N. et al. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N. Engl. J. Med. 370, 1889–1898 (2014).
Pawlotsky, J.M. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology 146, 1176–1192 (2014).
Jayasekera, C.R., Barry, M., Roberts, L.R. & Nguyen, M.H. Treating hepatitis C in lower-income countries. N. Engl. J. Med. 370, 1869–1871 (2014).
Pearlman, B.L. Protease inhibitors for the treatment of chronic hepatitis C genotype-1 infection: the new standard of care. Lancet Infect. Dis. 12, 717–728 (2012).
Schinazi, R., Halfon, P., Marcellin, P. & Asselah, T. HCV direct-acting antiviral agents: the best interferon-free combinations. Liver Int. 34 (Suppl. 1), 69–78 (2014).
Rauch, A. et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138, 1338–1345, 1345.e1–1345.e7 (2010).
Thomas, D.L. et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461, 798–801 (2009).
Ge, D. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401 (2009).
Suppiah, V. et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41, 1100–1104 (2009).
Tanaka, Y. et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat. Genet. 41, 1105–1109 (2009).
de Castellarnau, M. et al. Deciphering the interleukin 28B variants that better predict response to pegylated interferon-α and ribavirin therapy in HCV/HIV-1 coinfected patients. PLoS One 7, e31016 (2012).
di Iulio, J. et al. Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology 53, 1446–1454 (2011).
Pedergnana, V. et al. Analysis of IL28B variants in an Egyptian population defines the 20 kilobases minimal region involved in spontaneous clearance of hepatitis C virus. PLoS One 7, e38578 (2012).
Prokunina-Olsson, L. et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45, 164–171 (2013).
Sheahan, T. et al. Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe 15, 190–202 (2014).
Honda, M. et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 139, 499–509 (2010).
He, X.S. et al. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology 44, 352–359 (2006).
Welzel, T.M. et al. Variants in interferon-alpha pathway genes and response to pegylated interferon-Alpha2a plus ribavirin for treatment of chronic hepatitis C virus infection in the hepatitis C antiviral long-term treatment against cirrhosis trial. Hepatology 49, 1847–1858 (2009).
Fukuda, R. et al. Expression of interferon-alpha receptor mRNA in the liver in chronic liver diseases associated with hepatitis C virus: relation to effectiveness of interferon therapy. J. Gastroenterol. 31, 806–811 (1996).
Fukuda, R. et al. Effectiveness of interferon-alpha therapy in chronic hepatitis C is associated with the amount of interferon-alpha receptor mRNA in the liver. J. Hepatol. 26, 455–461 (1997).
Mathai, J. et al. IFN-alpha receptor mRNA expression in a United States sample with predominantly genotype 1a/I chronic hepatitis C liver biopsies correlates with response to IFN therapy. J. Interferon Cytokine Res. 19, 1011–1018 (1999).
Morita, K. et al. Expression of interferon receptor genes in the liver as a predictor of interferon response in patients with chronic hepatitis C. J. Med. Virol. 58, 359–365 (1999).
Morita, K. et al. Expression of interferon receptor genes (IFNAR1 and IFNAR2 mRNA) in the liver may predict outcome after interferon therapy in patients with chronic genotype 2a or 2b hepatitis C virus infection. J. Clin. Gastroenterol. 26, 135–140 (1998).
Liu, J. et al. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe 5, 72–83 (2009).
Chandra, P.K. et al. HCV infection selectively impairs type I but not type III IFN signaling. Am. J. Pathos. 184, 214–229 (2014).
Schwerk, J., Jarret, A.P., Joslyn, R.C. & Savan, R. Landscape of post-transcriptional gene regulation during hepatitis C virus infection. Curr. Opin. Virol. 12, 75–84 (2015).
van Rooij, E. et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 17, 662–673 (2009).
Liu, S. et al. Measuring antiviral activity of benzimidazole molecules that alter IRES RNA structure with an infectious hepatitis C virus chimera expressing Renilla luciferase. Antiviral Res. 89, 54–63 (2011).
Eldrup, A.B. et al. Structure-activity relationship of heterobase-modified 2′-C-methyl ribonucleosides as inhibitors of hepatitis C virus RNA replication. J. Med. Chem. 47, 5284–5297 (2004).
Ank, N. et al. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 80, 4501–4509 (2006).
Zeuzem, S. et al. Pegylated Interferon-Lambda (PEGIFN-L) shows superior viral response with improved safety and toleraility versus PEGIFNa-2A in HCV patients (G1/2/3/4): Emerage phase IIB through week 12. J. Hepatol. 54, S538–S539 (2011).
Muir, A. J. et al. A randomized phase 2b study of peginterferon lambda-1a for the treatment of chronic HCV infection. J. Hepatol. 61, 1238–1246 (2014).
Gonzalez, G., Pfannes, L., Brazas, R. & Striker, R. Selection of an optimal RNA transfection reagent and comparison to electroporation for the delivery of viral RNA. J. Virol. Methods 145, 14–21 (2007).
Acknowledgements
This project was funded partly by the Department of Immunology, Royalty Research Fund, University of Washington (R.S.) and by National Institutes of Health grants AI108765 (R.S.), AI060389, AI40035 (M.G.), and CA148068 (C.H.H.). This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E (M.C.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, and Center for Cancer Research. We thank S. Lemon and C. Rice for generously providing the H77D and JC1 viral strains, respectively; Y. Loo, S. Ozarkar and L. Kropp for experimental support; and C. Lim, A. Stone and the Gale and Savan labs for the discussions and feedback on the manuscript.
Author information
Authors and Affiliations
Contributions
A.J., A.P.M., and R.S. designed the study. A.J. analyzed the data and wrote the manuscript. R.S. directed the study. A.J., A.P.M., J.S., R.C.J. and M.H. performed experiments. A.J., S.M.H., A.K. and S.B. performed infections and flow cytometry preparations. D.P.B. provided reagents and reviewed the manuscript. D.P.B. was an employee of Biogen Idec and owns company stock. C.H.H., M.C. and M.G. provided intellectual input.
Corresponding author
Ethics declarations
Competing interests
Darren P Baker is an employee of Biogen Idec and an owner of Biogen Idec stock.
Supplementary information
Supplementary Text and Figures
Supplementary Texts 1–5 and Supplementary Figures 1–4 (PDF 2155 kb)
Rights and permissions
About this article
Cite this article
Jarret, A., McFarland, A., Horner, S. et al. Hepatitis-C-virus-induced microRNAs dampen interferon-mediated antiviral signaling. Nat Med 22, 1475–1481 (2016). https://doi.org/10.1038/nm.4211
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4211
This article is cited by
-
Direct and indirect effects of IFN-α2b in malignancy treatment: not only an archer but also an arrow
Biomarker Research (2022)
-
Revealing the characteristics of ZIKV infection through tissue-specific transcriptome sequencing analysis
BMC Genomics (2022)
-
microRNA-499a promotes the progression and chemoresistance of cervical cancer cells by targeting SOX6
Apoptosis (2020)
-
Differential expression of innate and adaptive immune genes in the survivors of three gibel carp gynogenetic clones after herpesvirus challenge
BMC Genomics (2019)
-
RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions
Nature Immunology (2019)