Abstract
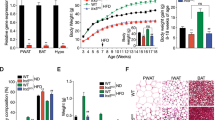
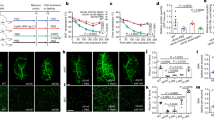
Adipocytes store excess energy in the form of triglycerides and signal the levels of stored energy to the brain. Here we show that adipocyte-specific deletion of Arntl (also known as Bmal1), a gene encoding a core molecular clock component, results in obesity in mice with a shift in the diurnal rhythm of food intake, a result that is not seen when the gene is disrupted in hepatocytes or pancreatic islets. Changes in the expression of hypothalamic neuropeptides that regulate appetite are consistent with feedback from the adipocyte to the central nervous system to time feeding behavior. Ablation of the adipocyte clock is associated with a reduced number of polyunsaturated fatty acids in adipocyte triglycerides. This difference between mutant and wild-type mice is reflected in the circulating concentrations of polyunsaturated fatty acids and nonesterified polyunsaturated fatty acids in hypothalamic neurons that regulate food intake. Thus, this study reveals a role for the adipocyte clock in the temporal organization of energy regulation, highlights timing as a modulator of the adipocyte-hypothalamic axis and shows the impact of timing of food intake on body weight.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Green, C.B., Takahashi, J.S. & Bass, J. The meter of metabolism. Cell 134, 728–742 (2008).
Reppert, S.M. & Weaver, D.R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002).
Marcheva, B. et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631 (2010).
Lamia, K.A., Storch, K.F. & Weitz, C.J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA 105, 15172–15177 (2008).
Oishi, K. et al. Disrupted fat absorption attenuates obesity induced by a high-fat diet in Clock mutant mice. FEBS Lett. 580, 127–130 (2006).
Turek, F.W. et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045 (2005).
Rudic, R.D. et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2, e377 (2004).
Ellingsen, T., Bener, A. & Gehani, A.A. Study of shift work and risk of coronary events. J. R. Soc. Promot. Health 127, 265–267 (2007).
Karlsson, B., Knutsson, A. & Lindahl, B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup. Environ. Med. 58, 747–752 (2001).
Spiegel, K., Tasali, E., Leproult, R. & Van Cauter, E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat. Rev. Endocrinol. 5, 253–261 (2009).
Ahima, R.S. et al. Role of leptin in the neuroendocrine response to fasting. Nature 382, 250–252 (1996).
Cowley, M.A. et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411, 480–484 (2001).
Mizuno, T.M. & Mobbs, C.V. Hypothalamic agouti-related protein messenger ribonucleic acid is inhibited by leptin and stimulated by fasting. Endocrinology 140, 814–817 (1999).
Lam, T.K. et al. Hypothalamic sensing of circulating fatty acids is required for glucose homeostasis. Nat. Med. 11, 320–327 (2005).
Pocai, A. et al. Restoration of hypothalamic lipid sensing normalizes energy and glucose homeostasis in overfed rats. J. Clin. Invest. 116, 1081–1091 (2006).
Cintra, D.E. et al. Unsaturated fatty acids revert diet-induced hypothalamic inflammation in obesity. PLoS ONE 7, e30571 (2012).
Bunger, M.K. et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 (2000).
Bunger, M.K. et al. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis 41, 122–132 (2005).
Kondratov, R.V., Kondratova, A.A., Gorbacheva, V.Y., Vykhovanets, O.V. & Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 20, 1868–1873 (2006).
Westgate, E.J. et al. Genetic components of the circadian clock regulate thrombogenesis in vivo. Circulation 117, 2087–2095 (2008).
He, W. et al. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA 100, 15712–15717 (2003).
Clausen, B.E., Burkhardt, C., Reith, W., Renkawitz, R. & Forster, I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 (1999).
Etchegaray, J.P., Lee, C., Wade, P.A. & Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182 (2003).
Ueda, H.R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 (2005).
Crumbley, C., Wang, Y., Kojetin, D.J. & Burris, T.P. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBα/RORα target gene. J. Biol. Chem. 285, 35386–35392 (2010).
Debruyne, J.P. et al. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50, 465–477 (2006).
Miller, B.H. et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. USA 104, 3342–3347 (2007).
Kornmann, B., Schaad, O., Bujard, H., Takahashi, J.S. & Schibler, U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5, e34 (2007).
Eguchi, J. et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13, 249–259 (2011).
Maffei, M. et al. Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat. Med. 1, 1155–1161 (1995).
El-Haschimi, K., Pierroz, D.D., Hileman, S.M., Bjorbaek, C. & Flier, J.S. Two defects contribute to hypothalamic leptin resistance in mice with diet-induced obesity. J. Clin. Invest. 105, 1827–1832 (2000).
Stunkard, A. et al. Binge eating disorder and the night-eating syndrome. Int. J. Obes. Relat. Metab. Disord. 20, 1–6 (1996).
Hogenesch, J.B., Gu, Y.Z., Jain, S. & Bradfield, C.A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 95, 5474–5479 (1998).
Oh, D.Y. et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698 (2010).
Wei, E. et al. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 11, 183–193 (2010).
Minokoshi, Y. et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428, 569–574 (2004).
Ropelle, E.R. et al. IL-6 and IL-10 anti-inflammatory activity links exercise to hypothalamic insulin and leptin sensitivity through IKKβ and ER stress inhibition. PLoS Biol. 8, e1000465 (2010).
Kiessling, S., Eichele, G. & Oster, H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J. Clin. Invest. 120, 2600–2609 (2010).
Pietiläinen, K.H. et al. Association of lipidome remodeling in the adipocyte membrane with acquired obesity in humans. PLoS Biol. 9, e1000623 (2011).
Arble, D.M., Bass, J., Laposky, A.D., Vitaterna, M.H. & Turek, F.W. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 17, 2100–2102 (2009).
Stucchi, P. et al. Circadian feeding drive of metabolic activity in adipose tissue and not hyperphagia triggers overweight in mice: is there a role of the pentose-phosphate pathway? Endocrinology 153, 690–699 (2012).
Hatori, M. et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012).
Masaki, T. et al. Involvement of hypothalamic histamine H1 receptor in the regulation of feeding rhythm and obesity. Diabetes 53, 2250–2260 (2004).
Salgado-Delgado, R., Angeles-Castellanos, M., Saderi, N., Buijs, R.M. & Escobar, C. Food intake during the normal activity phase prevents obesity and circadian desynchrony in a rat model of night work. Endocrinology 151, 1019–1029 (2010).
Fonken, L.K. et al. Light at night increases body mass by shifting the time of food intake. Proc. Natl. Acad. Sci. USA 107, 18664–18669 (2010).
Scheer, F.A., Hilton, M.F., Mantzoros, C.S. & Shea, S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. USA 106, 4453–4458 (2009).
Reyes, T.M., Walker, J.R., DeCino, C., Hogenesch, J.B. & Sawchenko, P.E. Categorically distinct acute stressors elicit dissimilar transcriptional profiles in the paraventricular nucleus of the hypothalamus. J. Neurosci. 23, 5607–5616 (2003).
Roberts, L.D. et al. Increased hepatic oxidative metabolism distinguishes the action of peroxisome proliferator-activated receptor δ from peroxisome proliferator-activated receptor γ in the ob/ob mouse. Genome Med. 1, 115 (2009).
Irizarry, R.A. et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 (2003).
Grant, G.R., Liu, J. & Stoeckert, C.J. Jr. A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics 21, 2684–2690 (2005).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grant RO1 HL097800 and Medical Research Council grant UD99999906. We thank M. Lazar for help with the ChIP experiments (funded by NIH R01 DK45586). We thank R. Ahima and the Mouse Phenotyping, Physiology and Metabolism Core of the Penn Diabetes Research Center (P30 DK19525) for performing body composition and behavioral analysis; D. Baldwin's Microarray Core Facility for performing the microarray analysis; and R. Freer and M. Adam for technical assistance. G.A.F. is the McNeil Professor of Translational Medicine and Therapeutics.
Author information
Authors and Affiliations
Contributions
G.K.P., S.I., W.-L.S., T.K., C.H.V. and F.W. contributed to the acquisition, analysis and interpretation of the data. G.K.P. and G.A.F. initiated and designed the study. G.G. performed statistical analyses and analysis of the microarray data. T.M.R. performed indirect calorimetric analyses. C.A.B. provided the mice with the conditional Arntl allele. M.E., M.M., J.L.G. and J.A.L. performed the liquid chromatography mass spectrometry (LC-MS) analysis. G.K.P. and G.A.F. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Methods (PDF 6800 kb)
Rights and permissions
About this article
Cite this article
Paschos, G., Ibrahim, S., Song, WL. et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med 18, 1768–1777 (2012). https://doi.org/10.1038/nm.2979
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2979
This article is cited by
-
Identification of the Relationship Between DNA Methylation of Circadian Rhythm Genes and Obesity
Biochemical Genetics (2024)
-
The evolving functions of the vasculature in regulating adipose tissue biology in health and obesity
Nature Reviews Endocrinology (2023)
-
Circadian mechanism disruption is associated with dysregulation of inflammatory and immune responses: a systematic review
Beni-Suef University Journal of Basic and Applied Sciences (2022)
-
The circadian clock influences T cell responses to vaccination by regulating dendritic cell antigen processing
Nature Communications (2022)
-
Associations between polymorphisms of SLC22A7, NGFR, ARNTL and PPP2R2B genes and Milk production traits in Chinese Holstein
BMC Genomic Data (2021)