Abstract
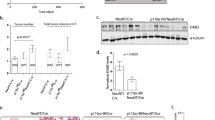
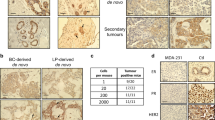
PIK3CA gain-of-function mutations are a common oncogenic event in human malignancy1,2,3,4, making phosphatidylinositol 3-kinase (PI3K) a target for cancer therapy. Despite the promise of targeted therapy, resistance often develops, leading to treatment failure. To elucidate mechanisms of resistance to PI3K-targeted therapy, we constructed a mouse model of breast cancer conditionally expressing human PIK3CAH1047R. Notably, most PIK3CAH1047R-driven mammary tumors recurred after PIK3CAH1047R inactivation. Genomic analyses of recurrent tumors revealed multiple lesions, including focal amplification of Met or Myc (also known as c-Met and c-Myc, respectively). Whereas Met amplification led to tumor survival dependent on activation of endogenous PI3K, tumors with Myc amplification became independent of the PI3K pathway. Functional analyses showed that Myc contributed to oncogene independence and resistance to PI3K inhibition. Notably, PIK3CA mutations and c-MYC elevation co-occur in a substantial fraction of human breast tumors. Together, these data suggest that c-MYC elevation represents a potential mechanism by which tumors develop resistance to current PI3K-targeted therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Samuels, Y. et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004).
Liu, P., Cheng, H., Roberts, T.M. & Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8, 627–644 (2009).
Vogt, P.K., Kang, S., Elsliger, M.A. & Gymnopoulos, M. Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem. Sci. 32, 342–349 (2007).
Yuan, T.L. & Cantley, L.C. PI3K pathway alterations in cancer: variations on a theme. Oncogene 27, 5497–5510 (2008).
Zhao, J.J. et al. The oncogenic properties of mutant p110α and p110β phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc. Natl. Acad. Sci. USA 102, 18443–18448 (2005).
Kang, S., Bader, A.G. & Vogt, P.K. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. USA 102, 802–807 (2005).
Isakoff, S.J. et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 65, 10992–11000 (2005).
Samuels, Y. et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 7, 561–573 (2005).
Engelman, J.A. et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 14, 1351–1356 (2008).
Gunther, E.J. et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 16, 283–292 (2002).
Raynaud, F.I. et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol. Cancer Ther. 8, 1725–1738 (2009).
O'Brien, C. et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3′ kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin. Cancer Res. 16, 3670–3683 (2010).
Engelman, J.A. et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007).
Turke, A.B. et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17, 77–88 (2010).
Zou, H.Y. et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 67, 4408–4417 (2007).
Sears, R. et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14, 2501–2514 (2000).
Yeh, E. et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6, 308–318 (2004).
Cizkova, M. et al. Gene expression profiling reveals new aspects of PIK3CA mutation in ERα-positive breast cancer: major implication of the Wnt signaling pathway. PLoS ONE 5, e15647 (2010).
Kan, Z. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010).
Haverty, P.M. et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosom. Cancer 47, 530–542 (2008).
Saal, L.H. et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc. Natl. Acad. Sci. USA 104, 7564–7569 (2007).
Stemke-Hale, K. et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN and AKT mutations in breast cancer. Cancer Res. 68, 6084–6091 (2008).
Tibes, R. et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 5, 2512–2521 (2006).
Zhao, X. et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 64, 3060–3071 (2004).
Moody, S.E. et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2, 451–461 (2002).
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006).
Moody, S.E. et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8, 197–209 (2005).
Tomayko, M.M. & Reynolds, C.P. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother. Pharmacol. 24, 148–154 (1989).
Acknowledgements
We thank T. Roberts, L. Cantley and W. Sellers for scientific discussions and suggestions. We thank L. Clayton and D. Silver for critical review of this manuscript. We thank R. Bronson for pathological analyses of tumor samples. We thank C. Li and E. Allgood for technical assistance. We thank L. Chodosh (University of Pennsylvania School of Medicine) for providing MMTV-rtTA mice. This work was supported by US National Institutes of Health grants CA134502 (J.J.Z.), CA148164-01 (J.J.Z. and N.S.G.) and K08CA122833 (R.B.), Stand Up To Cancer (J.J.Z. and G.B.M.), Dana Farber Harvard Cancer Center breast cancer SPORE grant P50 CA089393-08S1 (J.J.Z.), the US Department of Defense (BC051565 to J.J.Z.), the V Foundation (J.J.Z. and R.B.) and the Claudia Barr Program (J.J.Z.). In compliance with Harvard Medical School guidelines, we disclose that J.J.Z. and R.B. are consultants for Novartis Pharmaceuticals.
Author information
Authors and Affiliations
Contributions
P.L., H.C. and J.J.Z. designed the experiments, interpreted the data and wrote the paper. P.L. and H.C. carried out most of the experiments. S.S., A.I. and D.J.S. assisted with biochemical analyses and mouse work. J.Y., C.C., E.A.F., J.M. and R.S. carried out genome-wide DNA copy number profiling. N.S.G. provided GDC-0941 inhibitor. M.R. and R.B. analyzed co-occurrence of PIK3CA mutation with c-MYC amplification and overexpression in human breast tumors. F.Z. and G.B.M. provided the reverse-phase protein array data on the co-occurrence of PIK3CA mutation with increased c-MYC expression in human breast tumors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16, Supplementary Tables 1–3 and Supplementary Methods (PDF 2601 kb)
Rights and permissions
About this article
Cite this article
Liu, P., Cheng, H., Santiago, S. et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway–dependent and PI3K pathway–independent mechanisms. Nat Med 17, 1116–1120 (2011). https://doi.org/10.1038/nm.2402
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2402
This article is cited by
-
Nanopore sequencing from liquid biopsy: analysis of copy number variations from cell-free DNA of lung cancer patients
Molecular Cancer (2021)
-
Synergism between the phosphatidylinositol 3-kinase p110β isoform inhibitor AZD6482 and the mixed lineage kinase 3 inhibitor URMC-099 on the blockade of glioblastoma cell motility and focal adhesion formation
Cancer Cell International (2021)
-
Aspirin has a better effect on PIK3CA mutant colorectal cancer cells by PI3K/Akt/Raptor pathway
Molecular Medicine (2020)
-
Preclinical evaluation of a novel triple-acting PIM/PI3K/mTOR inhibitor, IBL-302, in breast cancer
Oncogene (2020)
-
Adaptation and selection shape clonal evolution of tumors during residual disease and recurrence
Nature Communications (2020)