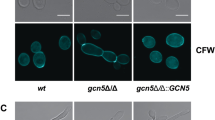
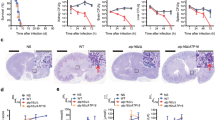
Abstract
Candida albicans is a major fungal pathogen that causes serious systemic and mucosal infections in immunocompromised individuals. In yeast, histone H3 Lys56 acetylation (H3K56ac) is an abundant modification regulated by enzymes that have fungal-specific properties, making them appealing targets for antifungal therapy. Here we demonstrate that H3K56ac in C. albicans is regulated by the RTT109 and HST3 genes, which respectively encode the H3K56 acetyltransferase (Rtt109p) and deacetylase (Hst3p). We show that reduced levels of H3K56ac sensitize C. albicans to genotoxic and antifungal agents. Inhibition of Hst3p activity by conditional gene repression or nicotinamide treatment results in a loss of cell viability associated with abnormal filamentous growth, histone degradation and gross aberrations in DNA staining. We show that genetic or pharmacological alterations in H3K56ac levels reduce virulence in a mouse model of C. albicans infection. Our results demonstrate that modulation of H3K56ac is a unique strategy for treatment of C. albicans and, possibly, other fungal infections.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Celic, I. et al. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 16, 1280–1289 (2006).
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle–regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005).
Hyland, E.M. et al. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 10060–10070 (2005).
Ozdemir, A. et al. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 280, 25949–25952 (2005).
Xu, F., Zhang, K. & Grunstein, M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121, 375–385 (2005).
Zhou, H., Madden, B.J., Muddiman, D.C. & Zhang, Z. Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry 45, 2852–2861 (2006).
Das, C., Lucia, M.S., Hansen, K.C. & Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459, 113–117 (2009).
Tjeertes, J.V., Miller, K.M. & Jackson, S.P. Screen for DNA-damage–responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 (2009).
Xie, W. et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol. Cell 33, 417–427 (2009).
Yuan, J., Pu, M., Zhang, Z. & Lou, Z. Histone H3–K56 acetylation is important for genomic stability in mammals. Cell Cycle 8, 1747–1753 (2009).
Driscoll, R., Hudson, A. & Jackson, S.P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315, 649–652 (2007).
Han, J. et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315, 653–655 (2007).
Tsubota, T. et al. Histone H3–K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25, 703–712 (2007).
Maas, N.L., Miller, K.M., DeFazio, L.G. & Toczyski, D.P. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23, 109–119 (2006).
Thaminy, S. et al. Hst3 is regulated by Mec1-dependent proteolysis and controls the S phase checkpoint and sister chromatid cohesion by deacetylating histone H3 at lysine 56. J. Biol. Chem. 282, 37805–37814 (2007).
Berman, J. & Sudbery, P.E. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3, 918–930 (2002).
Cowen, L.E., Anderson, J.B. & Kohn, L.M. Evolution of drug resistance in Candida albicans. Annu. Rev. Microbiol. 56, 139–165 (2002).
Pfaller, M.A. & Diekema, D.J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163 (2007).
Wisplinghoff, H. et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–317 (2004).
Sims, C.R., Ostrosky-Zeichner, L. & Rex, J.H. Invasive candidiasis in immunocompromised hospitalized patients. Arch. Med. Res. 36, 660–671 (2005).
Akins, R.A. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43, 285–318 (2005).
Perlin, D.S. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10, 121–130 (2007).
Wang, L., Tang, Y., Cole, P.A. & Marmorstein, R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18, 741–747 (2008).
Frye, R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 (2000).
Kelly, J., Rowan, R., McCann, M. & Kavanagh, K. Exposure to caspofungin activates Cap and Hog pathways in Candida albicans. Med. Mycol. 47, 697–706 (2009).
Roemer, T. et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50, 167–181 (2003).
Deere, D. et al. Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast 14, 147–160 (1998).
Sudbery, P., Gow, N. & Berman, J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12, 317–324 (2004).
Haldar, D. & Kamakaka, R.T. Schizosaccharomyces pombe Hst4 functions in DNA damage response by regulating histone H3 K56 acetylation. Eukaryot. Cell 7, 800–813 (2008).
Xhemalce, B. et al. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 282, 15040–15047 (2007).
Mullick, A. et al. Dysregulated inflammatory response to Candida albicans in a C5-deficient mouse strain. Infect. Immun. 72, 5868–5876 (2004).
Mullick, A. et al. Cardiac failure in C5-deficient A/J mice after Candida albicans infection. Infect. Immun. 74, 4439–4451 (2006).
Tuite, A., Elias, M., Picard, S., Mullick, A. & Gros, P. Genetic control of susceptibility to Candida albicans in susceptible A/J and resistant C57BL/6J mice. Genes Immun. 6, 672–682 (2005).
Maiese, K., Chong, Z.Z., Hou, J. & Shang, Y.C. The vitamin nicotinamide: translating nutrition into clinical care. Molecules 14, 3446–3485 (2009).
Dizdaroglu, M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res. 591, 45–59 (2005).
Bogan, K.L. & Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130 (2008).
Niren, N.M. Pharmacologic doses of nicotinamide in the treatment of inflammatory skin conditions: a review. Cutis 77, 11–16 (2006).
Cosgrove, M.S. et al. The structural basis of sirtuin substrate affinity. Biochemistry 45, 7511–7521 (2006).
Blander, G. et al. SIRT1 shows no substrate specificity in vitro. J. Biol. Chem. 280, 9780–9785 (2005).
Garske, A.L. & Denu, J.M. SIRT1 top 40 hits: use of one-bead, one-compound acetyl-peptide libraries and quantum dots to probe deacetylase specificity. Biochemistry 45, 94–101 (2006).
Saunders, L.R. & Verdin, E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene 26, 5489–5504 (2007).
Hirao, M. et al. Identification of selective inhibitors of NAD+-dependent deacetylases using phenotypic screens in yeast. J. Biol. Chem. 278, 52773–52782 (2003).
Pfaller, M.A. et al. Results from the ARTEMIS Drosoph. Inf. Serv.K global antifungal surveillance study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48, 1366–1377 (2010).
Pappas, P.G. et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 503–535 (2009).
Dagenais, T.R. & Keller, N.P. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22, 447–465 (2009).
Kushnirov, V.V. Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 (2000).
Gunjan, A. & Verreault, A.A. Rad53 kinase–dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115, 537–549 (2003).
Poveda, A. et al. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 279, 16033–16043 (2004).
Drogaris, P., Wurtele, H., Masumoto, H., Verreault, A. & Thibault, P. Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure-function relationships. Anal. Chem. 80, 6698–6707 (2008).
Schmitt, M.E., Brown, T.A. & Trumpower, B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18, 3091–3092 (1990).
Tsao, S., Rahkhoodaee, F. & Raymond, M. Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob. Agents Chemother. 53, 1344–1352 (2009).
Trunk, K. et al. Depletion of the cullin Cdc53p induces morphogenetic changes in Candida albicans. Eukaryot. Cell 8, 756–767 (2009).
Horsman, M.R., Siemann, D.W., Chaplin, D.J. & Overgaard, J. Nicotinamide as a radiosensitizer in tumours and normal tissues: the importance of drug dose and timing. Radiother. Oncol. 45, 167–174 (1997).
Acknowledgements
We are grateful to T. Roemer (Merck Research Laboratory) for providing strain CaSS1 and plasmids pHIS3 and pSAT-Tet, G. St-Germain (Laboratoire de santé publique du Québec) for strains MY067497, MY067743, MY070362, MY070589, MY069520, MY070954, MY070968 and MY070627, C. Restieri and M. Laverdière (Hôpital Maisonneuve-Rosemont) for strain 372.73374.1, C. Bachewich (Concordia University) for strain R153 and T. Edlind (Drexel University College of Medicine) for strain 20464. We are indebted to T. Roemer and M. Whiteway for critical reading of the manuscript. We thank F. Malenfant, J. Leroy, M. Mercier and P. Liscourt for excellent assistance in mouse handling. We are also grateful for the availability of the Candida Genome Database. This work was supported by research grants from the Canadian Institutes of Health Research to M.R. (CTP-79843, III-94587), A.M. (CTP-79843) and A.V. (CTP-79392) and from the National Science and Engineering Research Council of Canada to P.T. (STGP-322143-05). This is National Research Council of Canada publication number 52756. The Institute for Research in Immunology and Cancer is supported by funds from the Canadian Center of Excellence in Commercialization and Research, the Canadian Foundation for Innovation, and the Fonds de la Recherche en Santé du Québec.
Author information
Authors and Affiliations
Contributions
H.W. and S.T. designed and performed most experiments; G.L. constructed the yeast strains used in this study; J.T. and S.T. performed the fungal load and cytokine analyses; A.M. and M.R. designed and supervised the mouse studies; P.D., P.T. and A.V. designed and performed mass spectrometry experiments; E.-H.L. provided technical support; all authors participated in manuscript preparation; M.R. and A.V. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–4, Supplementary Results and Supplementary Methods (PDF 1105 kb)
Rights and permissions
About this article
Cite this article
Wurtele, H., Tsao, S., Lépine, G. et al. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat Med 16, 774–780 (2010). https://doi.org/10.1038/nm.2175
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.2175
This article is cited by
-
Old and new strategies in therapy and diagnosis against fungal infections
Applied Microbiology and Biotechnology (2024)
-
Multiple micronutrient deficiencies in early life cause multi-kingdom alterations in the gut microbiome and intrinsic antibiotic resistance genes in mice
Nature Microbiology (2023)
-
Morphogenic plasticity: the pathogenic attribute of Candida albicans
Current Genetics (2023)
-
Mechanisms of Candida Resistance to Antimycotics and Promising Ways to Overcome It: The Role of Probiotics
Probiotics and Antimicrobial Proteins (2021)
-
Lysine acetylation as drug target in fungi: an underexplored potential in Aspergillus spp.
Brazilian Journal of Microbiology (2020)