Green light from Jellyfish. © Marc Zimmer 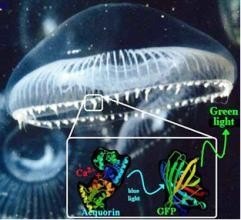
GFP, a 238 amino acids (26.9 kDa) protein, was first observed in the jellyfish Aequorea victoria occurring in partnership with the bioluminescent protein aequorin1. In this glowing jellyfish, GFP acts as a bioluminescence resonance energy transfer (BRET) acceptor, meaning it converts the blue emission of the bioluminescent protein (upon interaction with calcium ions) into a longer wavelength green emission. Possibly, the jellyfish uses this bioluminescence to attract secondary predators when attacked (the so-called 'burglar-alarm' hypothesis).
Albeit GFP was purified2 in 1962 by Osamu Shimomura, its primary amino acid sequence was only revealed3 in 1992 when Douglas Prasher opening the doors for its use as a tool in molecular biology. However, upon the loss of his funding he gave the cloned material to Martin Chalfie and left this field of science. Two years later4 Chalfie reported that the gene was self-sufficient to undergo the post-translational modifications necessary for chromophore formation by expressing it in the sensory neurons of the worm Caenorhabditis elegans as well as in the bacterium Escherichia coli. Shimomura's5 assignment of the chromophore structure was unambiguously confirmed by two independent reports of the X-ray crystal structure of GFP in 1996.
While Chalfie, Shimomura and Prasher were responsible for turning GFP into a tracer, it was Roger Tsien6 who elucidated the working principles of this molecule and dramatically improved its spectral characteristics resulting in increased fluorescence and photostability. Chalfie, Shimomura and Tsien shared the 2008 Nobel Prize in chemistry for their discovery and development of the green fluorescent protein while Prasher was given the noble snub as only three individuals can share a Nobel. Nevertheless this task could not have been accomplished without him. Like Chalfie said "(Prasher's) work was critical and essential for the work we did in our lab, they could've easily given the prize to Douglas and the other two and left me out."
The cytometric revolution
GFP has not only been a useful tool for monitoring protein expression and cellular events by microscopy, it has also played a significant role in flow cytometry. It is being used to identify and isolate cells from transgenic mice expressing proteins in unique cell lineages or at different stages of development, as also in the introduction of novel genes into cells. Many animals have been created to validate the concept that a gene can be expressed throughout a given organism. Using GFP as a marker a number of bacteria, yeast and other fungal cells, plant, fly, and mammalian (including human) cells have been created.
Prior to the advent of GFP, researchers could only analyse the distribution of proteins in a living cell by first fixing and thus killing the sample. No information could be collected from a protein within its natural environment. Even though it was easy to find where proteins are at a given time, it wasn't as easy to conceive how they got there. Thus, the most crucial piece in the puzzle remained elusive to cellular biologists across the world.
GFP and its derivatives that produce lighter and brighter versions of the protein covering the entire spectral range of the rainbow, have breathed a new life into fluorescence microscopy. The use of fluorescent proteins is less harmful as compared to chemicals like FITC (fluorescein isothiocyanate) in live cell imaging. This has led to a rapid development of wide range of live cell fluorescence microscopy systems over various time frames. These systems have advanced the understanding of numerous processes like neuronal development, cancer invasion and plaque accumulation in case of Alzheimer's disease.
"It's really been quite wonderful to see how many people have put their input into modifying, changing, and improving GFP so that it has become more and more useful every year." Chalfie, winner of 2008 Nobel Prize, said on being asked if he expected GFP to have this kind of impact.
The power of green
Marc Zimmer, professor at Connecticut College, whose lab has been working on GFP says, "I like to think of GFP as the microscope of the twenty-first century. Using GFP we can see when proteins are made, and where they can go."
This can be done by joining the GFP gene to the gene of the protein of interest so that when the protein is made it will have GFP hanging off it. Since GFP fluoresces, one can shine light at the cell and wait for the distinctive green fluorescence associated with GFP to appear.
By using DNA technology, researchers can now connect GFP to other interesting, but otherwise invisible proteins. This powerful marker allows them to track the movements, positions and interactions of the tagged proteins in a precise time and in physiological condition and locations. Some of the deepest mysteries of cellular biology have been explored and unravelled under the glowing green light of the GFP.
Jennifer Lippincort-Schwatz at the National Institute of Child Health and Human Development (NICHD), Bethesda has successfully used techniques like fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) to analyse specific movements of fluorescently tagged proteins within cells. These techniques, together with the relative ease of generating transgenic animals and the ability to express tagged proteins in specific tissues or at specific developmental times, provides a powerful means for the examination of behaviour of any tagged protein in embryos in myriad mutant backgrounds in vivo.
At Columbia University, labs of Martin Chalfie and Jonathan Javitch are inventing ways to use GFP as a tool to unravel several biological processes, particularly in the field of neuroscience. Chalfie's lab uses Caenorhabditis elegans to investigate aspects of nerve cell development, function and neuronal degeneration. Here, GFP is used as a gene and protein marker. Javitch's lab uses techniques like bioluminescent and fluorescent resonance energy transfer (BRET and FRET) spectroscopy methods to study protein-protein interaction for class A G-protein coupled receptors (GPCRs) by tagging the proteins with GFP derivatives. Structural and functional characterisation of this class of proteins is extremely important as they constitute one of the most important targets of the existing therapeutic arsenal.
Glowing in India
In India, several labs are involved in fascinating GFP research. For example, Amitabha Chattopadhyay's lab at Centre for Cellular and Molecular Biology, Hyderabad, uses GFP as a probe to monitor organization and dynamics of GPCRs such as the serotonin-1A receptor in healthy and diseased states. CCMB's Veena Parnaik along with Somdatta Sinha conceived a novel approach by tagging GFP for modeling gene circuits. She has also used GFP as a tag to study the localisation and dynamics of the splicing factor SC35.
"The impact of GFP on biological research in India has been tremendous like in the rest of the world," says Parnaik, CCMB Deputy Director.
At the International Centre for Genetic Engineering and Biotechnology, New Delhi, Virander Chauhan along with his colleagues Asif Mohmmed and Pawan Malhotra are involved in identification and functional characterisation of new drug/vaccine targets against malaria using a combination of molecular biological as well as functional genomics approaches. They use GFP fusion proteins to study targeting of parasite proteins, localisation in different stages and trafficking of parasite proteins within and outside the parasite.
M. N. Gupta's lab at IIT Delhi and several others at the Indian Institute of Science, Bangalore, have been using GFP in various fields of sciences.
PALM: Method of the year 2008
GFP has been rightly described as the microscope of the 21st century. The latest breakthrough in this field being PALM (the soothing acronym for photoactivated localisation microscopy): a super resolution microscope which allows for high resolution 2D and 3D images and single molecule tracking. The method was awarded the "Method of the year" award for 2008 by Nature Methods.
Green light from Jellyfish. © Marc Zimmer 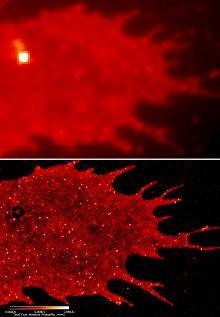
With PALM, they have been able to detect the precise cellular locations of thousands of individual protein molecule. With conventional fluorescence microscopy techniques, particularly at higher magnifications, these molecular masses appear as smeared fields of luminous blur. But PALM focuses those blurs, tremendously useful to biologists despite their lack of clarity, into amazing single points. All this is because of a single cleverly labelled protein.
As institutes invest time, money and efforts to explore innovative ways to use GFP as a tool to unravel challenging mysteries of cellular biology, it is only a matter of time before GFP is used to its full potential to reveal dynamic processes of macromolecular interactions in living cells.
GFP has had a lasting and dramatic impact on the field of biology. Be it cancer, neurological disorders or immunology, no single discipline has remained unaffected by the ability to visualise biological occurrences in real time in a physiological context.
The miracle of this kaleidoscope of colours remains to be fully exploited by scientists. "GFP has not outlived its usefulness. It has far more potential and it remains to be seen how people are going to use it." Chalfie says.
