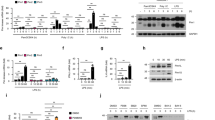
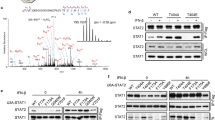
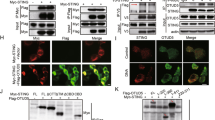
Abstract
Interferon (IFN) activates the signal transducer and activator of transcription (STAT) pathway to regulate immune responses. The protein inhibitor of activated STAT (PIAS) family has been suggested to negatively regulate STAT signaling. To understand the physiological function of PIAS1, we generated Pias1−/− mice. Using PIAS1-deficient cells, we show that PIAS1 selectively regulates a subset of IFN-γ- or IFN-β-inducible genes by interfering with the recruitment of STAT1 to the gene promoter. The antiviral activity of IFN-γ or IFN-β was consistently enhanced by Pias1 disruption. Pias1−/− mice showed increased protection against pathogenic infection. Our data indicate that PIAS1 is a physiologically important negative regulator of STAT1 and suggest that PIAS1 is critical for the IFN-γ- or IFN-β-mediated innate immune responses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Stark, G.R., Kerr, I.M., Williams, B.R., Silverman, R.H. & Schreiber, R.D. How cells respond to interferons. Annu. Rev. Biochem. 67, 227–264 (1998).
O'Shea, J.J., Gadina, M. & Schreiber, R.D. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109 (suppl.), S121–131 (2002).
Levy, D.E. & Darnell, J.E. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 (2002).
Shuai, K. & Liu, B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3, 900–911 (2003).
Alexander, W.S. Suppressors of cytokine signalling (SOCS) in the immune system. Nat. Rev. Immunol. 2, 410–416 (2002).
Hilton, D.J. Negative regulators of cytokine signal transduction. Cell. Mol. Life Sci. 55, 1568–1577 (1999).
ten Hoeve, J. et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 22, 5662–5668 (2002).
Liu, B. et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95, 10626–10631 (1998).
Chung, C.D. et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278, 1803–1805 (1997).
Arora, T. et al. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J. Biol. Chem. 278, 21327–21330 (2003).
Liu, B., Gross, M., ten Hoeve, J. & Shuai, K. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc. Natl. Acad. Sci. USA 98, 3203–3207 (2001).
Gil, M.P. et al. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. USA 98, 6680–6685 (2001).
Ramana, C.V. et al. Stat1-independent regulation of gene expression in response to IFN-γ. Proc. Natl. Acad. Sci. USA 98, 6674–6679 (2001).
Ramana, C.V., Gil, M.P., Schreiber, R.D. & Stark, G.R. Stat1-dependent and -independent pathways in IFN-γ-dependent signaling. Trends Immunol. 23, 96–101 (2002).
Maritano, D. et al. The STAT3 isoforms α and β have unique and specific functions. Nat. Immunol. 5, 401–409 (2004).
Kojima, H., Nakajima, K. & Hirano, T. IL-6-inducible complexes on an IL-6 response element of the junB promoter contain Stat3 and 36 kDa CRE-like site binding protein(s). Oncogene 12, 547–554 (1996).
Liu, K.D., Gaffen, S.L. & Goldsmith, M.A. JAK/STAT signaling by cytokine receptors. Curr. Opin. Immunol. 10, 271–278 (1998).
Pine, R., Canova, A. & Schindler, C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN α and IFN γ, and is likely to autoregulate the p91 gene. EMBO J. 13, 158–167 (1994).
Decker, T., Lew, D.J., Mirkovitch, J. & Darnell, J.E., Jr. Cytoplasmic activation of GAF, an IFN-γ-regulated DNA-binding factor. EMBO J. 10, 927–932 (1991).
Shuai, K., Schindler, C., Prezioso, V.R. & Darnell, J.E., Jr. Activation of transcription by IFN-γ: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258, 1808–1812 (1992).
Guyer, N.B., Severns, C.W., Wong, P., Feghali, C.A. & Wright, T.M. IFN-γ induces a p91/Stat1 a-related transcription factor with distinct activation and binding properties. J. Immunol. 155, 3472–3480 (1995).
Vinkemeier, U. et al. DNA binding of in vitro activated Stat1 a, Stat1 b and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 15, 5616–5626 (1996).
Khan, K.D. et al. Induction of the Ly-6A/E gene by interferon α/β and γ requires a DNA element to which a tyrosine-phosphorylated 91-kDa protein binds. Proc. Natl. Acad. Sci. USA 90, 6806–6810 (1993).
Wagner, B.J., Hayes, T.E., Hoban, C.J. & Cochran, B.H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 9, 4477–4484 (1990).
Varinou, L. et al. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-γ-dependent innate immunity. Immunity 19, 793–802 (2003).
Decker, T., Stockinger, S., Karaghiosoff, M., Muller, M. & Kovarik, P. interferons and STATs in innate immunity to microorganisms. J. Clin. Invest. 109, 1271–1277 (2002).
Anderson, S.L., Carton, J.M., Lou, J., Xing, L. & Rubin, B.Y. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology 256, 8–14 (1999).
Doyle, S. et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17, 251–263 (2002).
Martinez-Guzman, D. et al. Transcription program of murine gammaherpesvirus 68. J. Virol. 77, 10488–10503 (2003).
Meraz, M.A. et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84, 431–442 (1996).
Durbin, J.E., Hackenmiller, R., Simon, M.C. & Levy, D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450 (1996).
Car, B.D. et al. Interferon g receptor deficient mice are resistant to endotoxic shock. J. Exp. Med. 179, 1437–1444 (1994).
Karaghiosoff, M. et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat. Immunol. 4, 471–477 (2003).
Kinjyo, I. et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 17, 583–591 (2002).
Nakagawa, R. et al. SOCS-1 participates in negative regulation of LPS responses. Immunity 17, 677–687 (2002).
Kahyo, T., Nishida, T. & Yasuda, H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718 (2001).
Megidish, T., Xu, J.H. & Xu, C.W. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem. 277, 8255–8259 (2002).
Schmidt, D. & Muller, S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99, 2872–2877 (2002).
Johnson, E.S. & Gupta, A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 (2001).
Saitoh, H. & Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252–6258 (2000).
Rogers, R.S., Horvath, C.M. & Matunis, M.J. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J. Biol. Chem. 278, 30091–30097 (2003).
Ungureanu, D. et al. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood 102, 3311–3313 (2003).
Chin, A.I. et al. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416, 190–194 (2002).
Wu, T.T., Usherwood, E.J., Stewart, J.P., Nash, A.A. & Sun, R. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74, 3659–3667 (2000).
Acknowledgements
We thank R. Sun for antibodies to MHV-68 viral proteins; R. Schreiber for Jak1−/− cells; and J. Gao and B. Nguyen for technical assistance. Supported by the National Institutes of Health (K.S. and H.W.), American Cancer Society (K.S.) Howard Hughes Medical Institute (H.W.), Leukemia & Lymphoma Society (B.L.), National Institutes of Health–National Cancer Institute (training grant 5T32 CA009056 to S.M.), University of California at Los Angeles, (Sprague Jr. Fellowship to K.A.W.) and US Public Health Service (National Research Service award, GM07185 to K.A.W.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Tyrosine phosphorylation of Stat1 and Stat2 by IFN-γ or IFN-β. (PDF 104 kb)
Supplementary Table 1
Genes hyperactivated in Pias null BMMs in response to IFNs. (PDF 34 kb)
Supplementary Table 2
Primers used in real time quantitative PCR (Q-PCR) assays. (PDF 59 kb)
Rights and permissions
About this article
Cite this article
Liu, B., Mink, S., Wong, K. et al. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol 5, 891–898 (2004). https://doi.org/10.1038/ni1104
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1104
This article is cited by
-
Strawberry notch homolog 2 regulates the response to interleukin-6 in the central nervous system
Journal of Neuroinflammation (2022)
-
SENP1 prevents steatohepatitis by suppressing RIPK1-driven apoptosis and inflammation
Nature Communications (2022)
-
The JAK/STAT signaling pathway: from bench to clinic
Signal Transduction and Targeted Therapy (2021)
-
Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study
Signal Transduction and Targeted Therapy (2020)
-
Galectin-3 promotes Aβ oligomerization and Aβ toxicity in a mouse model of Alzheimer’s disease
Cell Death & Differentiation (2020)