Abstract
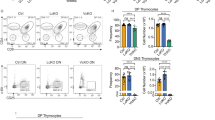
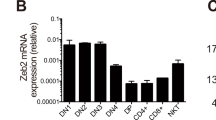
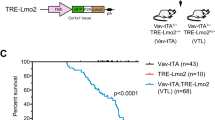
c-Myc is associated with cell growth and cycling in many tissues and its deregulated expression is causally implicated in cancer, particularly lymphomagenesis. However, the contribution of c-Myc to lymphocyte development is unresolved. We show here that the formation of normal lymphocytes by c-Myc−/− cells is selectively defective. c-Myc−/− cells are inefficient, in an age-dependent manner, at populating the thymus, and subsequent thymocyte maturation is ineffective: they fail to grow and proliferate normally at the late double-negative (DN) CD4−CD8− stage. Because N-Myc expression in thymocytes usually declines at the late DN stage, these results confirm that the nonredundant contributions of Myc family members to development are related to their distinct patterns of developmental gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Blackwood, E. M. & Eisenman, R. N. Max: A Helix-Loop-Helix Zipper Protein That Forms a Sequence-Specific DNA-Binding Complex with Myc. Science 251, 1211–1217 (1991).
Evan, G. I. & Littlewood, T. D. The role of c-myc in cell growth. Curr. Opin. Gen. Dev. 3, 44–49 (1993).
Dang, C. V. et al. Function of the c-Myc Oncogenic transcription Factor. Exp. Cell Res. 253, 63–77 (1999).
Malynn, B. A. et al. N-myc can functionally replace c-myc in murine development, cellular growth and differentiation. Genes Dev. 14, 1390–1399 (2000).
Berns, K., Hijmans, E. M., Koh, E., Daley, G. Q. & Bernards, R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene 19, 3330–3334 (2000).
Hatton, K. S. et al. Expression and Activity of L-Myc in Normal Mouse Development. Mol. Cell. Biol. 16, 1794–1804 (1996).
Charron, J. et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 6, 2248–2257 (1992).
Stanton, B. R., Perkins, A. S., Tessarollo, L., Sassoon, D. A. & Parada, L. F. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 6, 2235–2247 (1992).
Davis, A. C., Wims, M., Spotts, G. D., Hann, S. R. & Bradley, A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7, 671–682 (1993).
Sawai, S. et al. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development 117, 1445–1455 (1993).
Zimmerman, K. A. et al. Differential expression of myc family genes during murine development. Nature 319, 780–783 (1986).
Kelly, K., Cochran, B. H., Stiles, C. D. & Leder, P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell 35, 603–610 (1983).
Smith, R. K., Zimmerman, K., Yancopoulos, G. D., Ma, A. & Alt, F. W. Transcriptional Down-Regulation of N-myc Expression during B-Cell Development. Mol. Cell. Biol. 12, 1578–1584 (1992).
Miyazaki, T. Three Distinct IL-2 Signaling Pathways Mediated by bcl-2, c-myc and Lck Cooperate in Hematopoietic Cell Proliferation. Cell 81, 223–231 (1995).
Iritani, B. M. & Eisenman, R. N. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl Acad. Sci. USA 96, 13180–13185 (1999).
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985).
Leder, A., Pattengale, P. K., Kuo, A., Stewart, T. A. & Leder, P. Consequences of Widespread Deregulation of the c-myc Gene in Transgenic Mice: Multiple Neoplasms and Normal Development. Cell 45, 485–495 (1986).
Gardiner, E. M., Richman, A. & Hayday, A. Myc activation: a case of complex corruption. Semin. Virol. 2, 341–350 (1991).
Felsher, D. W. & Bishop, J. M. Reversible Tumorigenesis by MYC in Hematopoietic Lineages. Mol. Cell 4, 199–207 (1999).
Hoffman, E. S. et al. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 10, 948–962 (1996).
von Boehmer, H. & Fehling, H. J. Structure and function of the pre-T-cell receptor. Ann. Rev. Immunol. 15, 433–452 (1997).
Newton, K., Harris, A. W. & Strasser, A. FASS/MORTI regulates the pre-TCR checkpoint and can function as a tumour suppressor. EMBO J. 19, 931–40 (2000).
Pearson, R. & Weston, K. c-Myb regulates the proliferation of immature thymocytes following β-selection. EMBO J. 19, 6112–6120 (2000).
Okamura, R. M. et al. Redundant Regulation of T Cell Differentiation and TCRα Gene Expression by the Transcription Factors LEF-1 and TCF-1. Immunity 8, 11–20 (1998).
Colcucci, F. A new look at syk in αβ and γδ T cell development using chimeric mice with a low competitive hematopoietic environment. J. Immunol. 164, 5140–5145 (2000).
Mallick-Wood, C. A. et al. Disruption of epithelial γδ T cell repertoires by mutation of the Syk tyrosine kinase. Proc. Natl Acad. Sci. USA 93, 9704–9709 (1996).
Mombaerts, P. et al. RAG-1-Deficient Mice Have No Mature B and T Lymphocytes. Cell 68, 869–877 (1992).
Lefrancois, L. Extrathymic differentiation of intraepithelial lymphocytes, generation of a separate and unequal T-cell repertoire. Immunol. Today 12, 436–438 (1991).
Klein, M. Jr Peripheral engraftment of fetal intestine into athymic mice sponsors T-cell development: direct evidence for thymopoietic function of murine small intestine. J. Exp. Med. 176, 1365–1373 (1992).
Mombaerts, P. et al. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature 360, 225–231 (1992).
Mallick, C. A., Dudley, E. C., Viney, J. L., Owen, M. J. & Hayday, A. C. Rearrangement and Diversity of T Cell Receptor β Chain Genes in Thymocytes: A Critical Role for the β Chain in Development. Cell 73, 513–519 (1993).
Fehling, H. J., Krotkova, A., Saint-Ruf, C. & von Boehmer, H. Crucial role of the pre-T-cell receptor α gene in development of αβ but not γδ T cells. Nature 375, 795–798 (1995).
Shores, E. W., Sharrow, S. O., Uppenkamp, I. & Singer, A. T cell receptor-negative thymocytes from SCID mice can be induced to enter the CD4/CD8 differentiation pathway. Eur. J. Immunol. 20, 69–77 (1990).
Ikuta, K. et al. A developmental Switch in Thymic Lymphocyte Maturation Potential Occurs at the Level of Hematopoietic Stem Cells. Cell 62, 863–874 (1990).
Dudley, E. C., Petrie, H. T., Shah, L. M., Owen, M. J. & Hayday, A. C. T cell receptor β chain gene rearrangement and selection during thymocyte development in adult mice. Immunity 1, 83–93 (1994).
Broussard-Diehl, C., Bauer, S. R. & Scheuermann, R. H. A Role for c-myc in the Regulation of Thymocyte Differentiation and Possibly Positive Selection. J. Immunol. 156, 3141–3150 (1996).
Alt, F. W. The human myc gene family. Cold Spring Harb. Symp. Quant. Biol. 51, 931–941 (1986).
Lemischka, I. R., Raulet, D. H. & Mulligan, R. C. Development Potential and Dynamic Behavior of Hematopoietic Stem Cells. Cell 45, 917–927 (1986).
Wang, J., Xie, S., Allan, D., Beach, D. & Hannon, G. J. Myc activates telomerase. Genes Dev. 12, 1769–1774 (1998).
Blasco, M. A. & et al. telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 (1997).
Moreno de Alboran, I. M. et al. Analysis of c-Myc function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55 (2001).
Passoni, L. et al. Intrathymic δ Selection Events in γδ Cell Development. Immunity 7, 83–95 (1997).
Johnston, L. A., Prober, D. A., Edgar, B. A., Eisenman, R. N. & Gallant, P. Drosophila myc Regulates Cellular Growth during Development. Cell 98, 779–790 (1999).
Vlach, J., Hennecke, S., Alevizopouolos, K., Conti, D. & Amati, B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 15, 6595–6604 (1996).
von Boehmer, H. et al. Crucial function of the pre-T-cell receptor (TCR) in TCRβ selection, TCR β allelic exclusion and αβ versus γδ lineage commitment. Immunol. Rev. 165, 111–119 (1998).
O'Hagan, R. C. et al. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 14, 2185–2191 (2000).
Voll, R. E. et al. NF-κB activation by the pre-T cell receptor serves as a selective survival signal in T-lymphocyte development. Immunity 13, 677–689 (2000).
Seckinger, P., Milili, M., Schiff, C. & Fourgereau, M. Interleukin-7 regulates c-myc expression in murine T cells and thymocytes: a role for tyrosine kinase(s) and calcium mobilization. Eur. J. Immunol. 24, 716–722 (1994).
Morrow, M. A., Lee, G., Gillis, S., Yancopoulos, G. D. & Alt, F. W. Interleukin-7 induces N-myc and c-myc expression in normal precursor B lymphocytes. Genes Dev. 6, 61–70 (1992).
von-Freeden-Jeffry, U. et al. Lymphopenia in Interleukin (IL)-7 Gene-deleted Mice Identifies IL-7 as a Nonredundant Cytokine. J. Exp. Med. 1995, 1519–1526 (1995).
Peschon, J. J. et al. Early Lymphocyte Expansion Is Severely Impaired in Interleukin 7 Receptor-deficient Mice. J. Exp. Med. 180, 1955–1960 (1994).
Maki, K. et al. Interleukin 7 receptor-deficient mice lack γδ T cells. Proc. Natl Acad. Sci. USA 93, 7172–7177 (1996).
Papaioannou, V. & Johnson, R. in Gene Targeting: A practical approach (eds Rickwood, D. & Hames, B.D.) 107–146 (Oxford University Press, New York, 1993).
Chen, J., Lansford, R., Stewart, V., Young, F. & Alt, F. W. RAG-2-deficient blastocyst complementation: An assay of gene function in lymphocyte development. Proc. Natl Acad. Sci. USA 90, 4528–4532 (1993).
Dudley, E. C., Girardi, M., Owen, M. J. & Hayday, A. C. αβ and γδ T cells can share a late common precursor. Curr. Biol. 5, 659–669 (1995).
Scheuermann, R. H. & Bauer, S. R. Polymerase Chain Reaction-Based mRNA Quantification Using An Internal Standard: Analysis of Oncogene Expression. Meth. Enzymol. 218, 446–473 (1993).
Acknowledgements
We thank D. Barber, R. Carbone, J. Cridland, S. Maher, J. McGrath, D. Schatz, T. Taylor, R. Tigelaar, W. Turnbull, E. Hoffman, L. Passoni, S. John, S. Roberts, M. Owen, J. Lewis, S. Creighton and, particularly, R. Sullo. Supported by the Wellcome Trust and by National Institutes of Health grant GM37759 (to A. C. H.). N. C. D. is supported by MSTP.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Web Table 1.
TCRβ and TCRδ gene rearrangements in thymocytes of c-Myc−/− RAG-1−/− chimeras (DOC 32 kb)
Web Table 2.
TCRγ gene rearrangements in thymocytes of c-Myc−/−RAG-1−/− chimeras, arranged according to germline contributions from V and J segments and presumed template-independent N or P nucleotides (DOC 21 kb)
Web Table 3.
The frequencies of in-frame TCRβ gene rearrangements (DOC 19 kb)
Rights and permissions
About this article
Cite this article
Douglas, N., Jacobs, H., Bothwell, A. et al. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol 2, 307–315 (2001). https://doi.org/10.1038/86308
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/86308
This article is cited by
-
MNT suppresses T cell apoptosis via BIM and is critical for T lymphomagenesis
Cell Death & Differentiation (2023)
-
Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat
Signal Transduction and Targeted Therapy (2020)
-
GATA3 induces human T-cell commitment by restraining Notch activity and repressing NK-cell fate
Nature Communications (2016)
-
Transcription factors and target genes of pre-TCR signaling
Cellular and Molecular Life Sciences (2015)
-
Distinct TCR signaling pathways drive proliferation and cytokine production in T cells
Nature Immunology (2013)