Abstract
SHARPIN forms a linear-ubiquitin-chain–assembly complex that promotes signaling via the transcription factor NF-κB. SHARPIN deficiency leads to progressive multi-organ inflammation and immune system malfunction, but how SHARPIN regulates T cell responses is unclear. Here we found that SHARPIN deficiency resulted in a substantial reduction in the number of and defective function of regulatory T cells (Treg cells). Transfer of SHARPIN-sufficient Treg cells into SHARPIN-deficient mice considerably alleviated their systemic inflammation. SHARPIN-deficient T cells displayed enhanced proximal signaling via the T cell antigen receptor (TCR) without an effect on the activation of NF-κB. SHARPIN conjugated with Lys63 (K63)-linked ubiquitin chains, which led to inhibition of the association of TCRζ with the signaling kinase Zap70; this affected the generation of Treg cells. Our study therefore identifies a role for SHARPIN in TCR signaling whereby it maintains immunological homeostasis and tolerance by regulating Treg cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Ikeda, F. & Dikic, I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 9, 536–542 (2008).
Pickart, C.M. & Fushman, D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 (2004).
Lim, S. et al. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol. Cell. Neurosci. 17, 385–397 (2001).
Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 (2011).
Tokunaga, F. et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011).
HogenEsch, H. et al. A spontaneous mutation characterized by chronic proliferative dermatitis in C57BL mice. Am. J. Pathol. 143, 972–982 (1993).
Seymour, R.E. et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 8, 416–421 (2007).
Liang, Y., Seymour, R.E. & Sundberg, J.P. Inhibition of NF-κB signaling retards eosinophilic dermatitis in SHARPIN-deficient mice. J. Invest. Dermatol. 131, 141–149 (2011).
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 (2009).
Pouwels, J. et al. SHARPIN regulates uropod detachment in migrating lymphocytes. Cell Rep. 5, 619–628 (2013).
Rantala, J.K. et al. SHARPIN is an endogenous inhibitor of β1-integrin activation. Nat. Cell Biol. 13, 1315–1324 (2011).
Potter, C.S. et al. Chronic proliferative dermatitis in Sharpin null mice: development of an autoinflammatory disease in the absence of B and T lymphocytes and IL4/IL13 signaling. PLoS ONE 9, e85666 (2014).
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Fontenot, J.D., Gavin, M.A. & Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Bennett, C.L. et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 (2001).
Brunkow, M.E. et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27, 68–73 (2001).
Ohkura, N. & Sakaguchi, S. Regulatory T cells: roles of T cell receptor for their development and function. Semin. Immunopathol. 32, 95–106 (2010).
Brownlie, R.J. & Zamoyska, R. T cell receptor signalling networks: branched, diversified and bounded. Nat. Rev. Immunol. 13, 257–269 (2013).
Chuck, M.I., Zhu, M., Shen, S. & Zhang, W. The role of the LAT-PLC-γ1 interaction in T regulatory cell function. J. Immunol. 184, 2476–2486 (2010).
Koonpaew, S., Shen, S., Flowers, L. & Zhang, W. LAT-mediated signaling in CD4+CD25+ regulatory T cell development. J. Exp. Med. 203, 119–129 (2006).
Siggs, O.M. et al. Opposing functions of the T cell receptor kinase ZAP-70 in immunity and tolerance differentially titrate in response to nucleotide substitutions. Immunity 27, 912–926 (2007).
Tanaka, S. et al. Graded attenuation of TCR signaling elicits distinct autoimmune diseases by altering thymic T cell selection and regulatory T cell function. J. Immunol. 185, 2295–2305 (2010).
Hwang, S. et al. Reduced TCR signaling potential impairs negative selection but does not result in autoimmune disease. J. Exp. Med. 209, 1781–1795 (2012).
Wang, Z., Sokolovska, A., Seymour, R., Sundberg, J.P. & Hogenesch, H. SHARPIN is essential for cytokine production, NF-κB signaling, and induction of Th1 differentiation by dendritic cells. PLoS ONE 7, e31809 (2012).
Harada, Y. et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 207, 1381–1391 (2010).
Josefowicz, S.Z., Lu, L.F. & Rudensky, A.Y. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012).
Janssens, S., Tinel, A., Lippens, S. & Tschopp, J. PIDD mediates NF-κB activation in response to DNA damage. Cell 123, 1079–1092 (2005).
Huang, H. et al. K33-linked polyubiquitination of T cell receptor-ζ regulates proteolysis-independent T cell signaling. Immunity 33, 60–70 (2010).
Jin, H.S., Liao, L., Park, Y. & Liu, Y.C. Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc. Natl. Acad. Sci. USA 110, 624–629 (2013).
Kulathu, Y. & Komander, D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 (2012).
Wang, H. et al. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb. Perspect. Biol. 2, a002279 (2010).
Liang, Y. & Sundberg, J.P. SHARPIN regulates mitochondria-dependent apoptosis in keratinocytes. J. Dermatol. Sci. 63, 148–153 (2011).
HogenEsch, H., Torregrosa, S.E., Boggess, D., Sundberg, B.A., Carroll, J. & Sundberg, J.P. Increased expression of type 2 cytokines in chronic proliferative dermatitis (cpdm) mutant mice and resolution of inflammation following treatment with IL-12. Eur. J. Immunol. 31, 734–742 (2001).
Renninger, M.L., Seymour, R.E., Whiteley, L.O., Sundberg, J.P. & Hogenesch, H. Anti-IL5 decreases the number of eosinophils but not the severity of dermatitis in Sharpin-deficient mice. Exp. Dermatol. 19, 252–258 (2010).
Wang, Z., Potter, C.S., Sundberg, J.P. & Hogenesch, H. SHARPIN is a key regulator of immune and inflammatory responses. J. Cell. Mol. Med. 16, 2271–2279 (2012).
Levine, A.G., Arvey, A., Jin, W. & Rudensky, A.Y. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014).
Holst, J. et al. Scalable signaling mediated by T cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nat. Immunol. 9, 658–666 (2008).
Huehn, J., Polansky, J.K. & Hamann, A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat. Rev. Immunol. 9, 83–89 (2009).
Sakaguchi, N. et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature 426, 454–460 (2003).
Gomez-Rodriguez, J. et al. Itk-mediated integration of T cell receptor and cytokine signaling regulates the balance between Th17 and regulatory T cells. J. Exp. Med. 211, 529–543 (2014).
Sauer, S. et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc. Natl. Acad. Sci. USA 105, 7797–7802 (2008).
van Oers, N.S., Killeen, N. & Weiss, A. ZAP-70 is constitutively associated with tyrosine-phosphorylated TCRζ in murine thymocytes and lymph node T cells. Immunity 1, 675–685 (1994).
Acknowledgements
We thank A. Rudensky (Memorial Sloan Kettering Cancer Center) for Foxp3YFPCre mice. Supported by the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (RO1AI62969, RO1AI78272 and PO1AI089624).
Author information
Authors and Affiliations
Contributions
Y.P. and H.J. designed and performed the experiments, analyzed the data, and wrote the manuscript; J.Lo. and C.E. did the mouse breeding and helped with the preparation of experiments; J.Le. helped with experimental design and data analysis; L.L. performed the proteomics analysis; and Y.-C.L. initiated and secured funding for this project, helped with experimental design and data interpretation, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
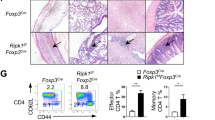
Supplementary Figure 1 Phenotypic analysis of thymocytes.
(a) Expression level of SHARPIN in DN, CD4+, CD8+ SP and DP thymocytes, CD4+ naïve T and Treg cells, B cells and macrophages. (b) Total numbers of CD4+ SP, CD8+ SP and DP thymocytes of Cpdm+/− and Cpdm−/− mice (n= 8 mice per group). (c) Flow cytometric analysis (left) or frequencies (right) of CD4+ and CD8+ T cells in the thymus of 4 wk old Cpdm+/− and Cpdm−/− mice (n= 10 mice per group). (d) Percentage of Live/Dead+ DP thymocytes from Cpdm+/− and Cpdm−/− mice with or without anti-CD3 and anti-CD28 stimulation (n= 8 mice per group). The data are compiled or representative of three to four independent experiments. Error bars indicate the mean (± s.d.). *P < 0.05 (two-tailed unpaired t-test).
Supplementary Figure 2 Analysis of T cell development and phenotype in BM chimeric mice.
(a-b) Flow cytometric analysis of CD4+ and CD8+ in pregated CD45.1+ or CD45.2+ cells of Thy or SP from mice reconstituted with individual (a) or mixed transfers (b) of WT or Cpdm+/− and Cpdm−/− BM (n= 4-6 mice per group). (c) Sorted naive CD4+CD62L+CD44−CD25− T cells of SP from mice reconstituted with mixed transfers of WT or Cpdm+/− and Cpdm−/− BM were cultured with anti-CD3 or anti-CD3 and anti-CD28. Cell Trace Violet dilution was examined 4 d later by flow cytometry. (d) Percentage of Live/Dead+ CD4+ T cells of SP from the mice in (c) with or without anti-CD3 and anti-CD28 stimulation. (e) Flow cytometric analysis (upper) or frequencies (bottom) of CD62L+ and CD44+ of CD4+ T cells in SP or lung from mice reconstituted with individual or mixed transfers of WT or Cpdm+/− and Cpdm−/− BM (n= 6-8 mice per group). (f) Cytokine production in CD4+T cells. Isolated lung cells from the mice in (e) were stimulated with PMA and Ionomycin for 4h (n= 6 mice per group). (g) H&E staining (upper) and histology scores (bottom) of the lung tissue section from the mice in (e). Original magnification, ×100 (n= 6 mice per group). (h) Analysis of the phosphorylation status of signaling proteins in sorted CD4+ T cells from Cpdm+/− and Cpdm−/− mice upon TGF-β stimulation. The data are compiled or representative of two to three independent experiments. Error bars indicate the mean (± s.d.). *P < 0.05 **P < 0.01 and ***P < 0.001 (two-tailed unpaired t-test).
Supplementary Figure 3 Analysis of Treg cell proliferation and apoptosis.
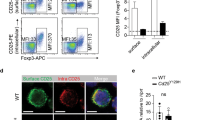
(a) Flow cytometric analysis of Ki-67 expression in Treg cells of SP from Cpdm+/− and Cpdm−/− mice (n= 12 mice per group). (b) Sorted CD4+CD25+Nrp1+ Treg cells of SP from Cpdm+/− and Cpdm−/− mice were cultured with anti-CD3and anti-CD28. Cell Trace Violet dilution was examined 2 or 4 d later by flow cytometry. (c) Percentage of Live/Dead+ CD4+Foxp3+ Treg cells with or without anti-CD3 and anti-CD28 in SP from Cpdm+/− and Cpdm−/− mice (n= 8 mice per group). (d) Flow cytometric analysis of Ki-67 expression in Treg cells of SP from mixed chimeric mice (n= 8 mice per group). (e) Percentage of Live/Dead+ CD4+Foxp3+ Treg cells with or without anti-CD3 and anti-CD28 in SP from mixed chimeric mice (n= 6 mice per group). (f-g) Percentage of Live/Dead+ CD4+Foxp3+ Treg cells in thymus from Cpdm+/− and Cpdm−/− mice (f) or mixed chimeric mice (g) (n= 6 mice per group). The data are compiled or representative of two to four independent experiments. Error bars indicate the mean (± s.d.). *P < 0.05 **P < 0.01 and ***P < 0.001 (two-tailed unpaired t-test).
Supplementary Figure 4 SHARPIN-deficient Treg cells have a defect in the suppression of colitis.
Rag1−/− mice received sorted CD4+CD25+YFP+ Treg (CD45.2+) cells from Foxp3YFPCreCpdm+/−or Foxp3YFPCreCpdm−/− mice, together with CD4+CD45RBhi (CD45.1+) T cells, or CD4+CD45RBhi (CD45.1+) T cells alone (None). (a-d) Colon length (a), the ratios of CD4+CD45RBhigh (CD45.1+) to CD4+YFP+ (CD45.2+) Treg cells in the indicated organs (b), absolute numbers of CD4+YFP+ (CD45.2+) Treg cells in the indicated organs (c), and frequencies of the indicated cytokine-producing CD4+CD45RBhi (CD45.1+) T cells in the indicated organs (d) of the Rag1−/− recipient mice at 8 wk after transfer (n= 6 to 12 mice per group). (e-f) Isolated CD4+ T cells from Cpdm+/− and Cpdm−/− mice were differentially labeled with Cell Trace Far Red DDAO-SE and Violet dye, respectively, or vice versa, and then co-injected into WT recipient mice. Flow cytometric analysis (left) or percentage (right) of the input population and transferred CD4+Foxp3− T cells (e), flow cytometric analysis (left) or percentage (right) of the input population and transferred CD4+Foxp3+ T cells (f), isolated from various tissues of the recipient mice at 2 or 20 hr after injection (n= 6 mice per group). The data are compiled or representative of two to four independent experiments. Error bars indicate the mean (± s.d.). *P < 0.05 **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired t-test).
Supplementary Figure 5 Cytokine production in SHARPIN-deficient memory or naive T cells or Treg cells.
(a) The methylation pattern of CNS2 of the Foxp3 locus in sorted CD4+YFP+CD25+ Treg cells and CD4+YFP−CD25− conventional T cells from Foxp3YFPCreCpdm+/−or Foxp3YFPCreCpdm−/− mice. Individual CpG motif within CNS2 is vertically arranged and color-coded according to the degree of methylation at that site (n= 6 mice per group). (b) Sorted CD45.1+CD4+CD25+Nrp1+ or CD45.2+CD4+CD25+Nrp1+ Treg cells from mixed chimeric mice were cultured with anti-CD3 and anti-CD28 for 36 h. Cytokine production was measured by bio-plex multi-cytokine assay (n= 6 mice per group). (c) Flow cytometric analysis of Foxp3 (upper) or RORγt (bottom) expression in CD4+ T cells from Cpdm+/− and Cpdm−/− mice upon stimulation with the indicated concentration of anti-CD3 or anti-CD3 and anti-CD28 in the presence of TGF-β (1ng/ml) and IL-2. (d-e) Cytokine production was measured by bio-plex multi-cytokine assay in sorted memory CD4+CD44+CD62L− T cells (d) or naïve CD4+CD62L+CD44−CD25− T cells (e) from Cpdm+/− and Cpdm−/− mice upon TCR and CD28 stimulation for 36 h. f, Sorted naïve CD4+CD62L+CD44−CD25− T cells from Cpdm+/− and Cpdm−/− mice were cultured under TH0, TH1, TH2 and TH17 polarizing conditions for 5 days. IFN-γ, IL-4 or IL-17A producing cells were analyzed by intracellular cytokine staining and flow cytometry 6 h after restimulation with anti-CD3 and anti-CD28 (n= 6 mice per group). The data are compiled or representative of two to four independent experiments. Error bars indicate the mean (± s.d.). *P < 0.05 **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired t-test).
Supplementary Figure 6 Phosphorylation status of signaling proteins upon stimulation with various stimuli.
(a) Analysis of the phosphorylation status of signaling proteins in sorted CD4+ T cells from Cpdm+/− and Cpdm−/− mice upon TCR and CD28 stimulation. (b) Analysis of the phosphorylation status of signaling proteins in sorted DP thymocytes from Cpdm+/− and Cpdm−/− mice upon TCR stimulation. (c) Analysis of the phosphorylation status of signaling proteins in sorted CD4+ T cells from Cpdm+/− and Cpdm−/− mice upon TNF stimulation. (d) Splenocytes from Cpdm+/− and Cpdm−/− mice were stimulated with PMA and Ionomycin for 10 min. Phosphorylation status of ERK1/2 in CD4+Foxp3− T or CD4+Foxp3+ Treg cells were analyzed by intracellular phosphor-protein staining. The data are representative of three independent experiments.
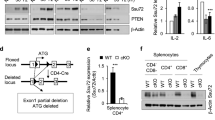
Supplementary Figure 7 Linkage-specific ubiquitination and the mapping of ubiquitination sites on SHARPIN.
(a-c) Jurkat T cells were transiently expressed with the indicated plasmids and analyzed for ubiquitination. (a) HA-Ub WT and SHARPIN WT or mutants (F354V, K42R, K168/169R and K312R); (b) HA-Ub WT or mutants (K11, K48 and K63) and SHARPIN WT or mutants (K42R, K168/169R and K312R); and (c) HA-Ub WT or mutants (K11 and K48) and SHARPIN WT or mutants (F354V and K312R). Cells were stimulated with OKT3 for 5 min, and lysed by adding SDS lysis buffer (2% SDS) and immunoprecipitated with anti-Myc antibody. The data are representative of three independent experiments.
Supplementary Figure 8 Knockdown of SHARPIN reduces the generation of Treg cells, but knockdown of HOIP does not.
(a) Analysis of mAmetrine expression in peripheral blood (left) and SHARPIN or HOIP protein expression (right) from BM chimeric mice 8 wks after reconstitution with retrovirus vector expressing control, HOIP or SHARPIN shRNA. (b) Splenocytes from WT mice were stimulated with concanavalin A, followed by retroviral transduction of retrovirus vector expressing control, HOIP or SHARPIN shRNA. Four days after transduction, the phosphorylation status of signaling proteins was analyzed in sorted mAmetrine+CD4+T cells upon TCR stimulation. (c-d) Flow cytometric analysis of Foxp3 expression in pregated mAmetrine+CD45.2+CD4+ T cells in thymus (c) or SP (d) of the mice in (a) (n= 6 mice per group). The data are compiled or representative of three independent experiments with two mice each. Error bars indicate the mean (± s.d.). *P < 0.05 **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired t-test).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 2 (PDF 2854 kb)
Rights and permissions
About this article
Cite this article
Park, Y., Jin, Hs., Lopez, J. et al. SHARPIN controls regulatory T cells by negatively modulating the T cell antigen receptor complex. Nat Immunol 17, 286–296 (2016). https://doi.org/10.1038/ni.3352
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3352
This article is cited by
-
SHARPIN: Role in Finding NEMO and in Amyloid-Beta Clearance and Degradation (ABCD) Pathway in Alzheimer’s Disease?
Cellular and Molecular Neurobiology (2022)
-
Met1-linked ubiquitin signalling in health and disease: inflammation, immunity, cancer, and beyond
Cell Death & Differentiation (2021)
-
Dual roles for LUBAC signaling in thymic epithelial cell development and survival
Cell Death & Differentiation (2021)
-
Integrative proteomics reveals an increase in non-degradative ubiquitylation in activated CD4+ T cells
Nature Immunology (2019)
-
Modulation of autoimmune pathogenesis by T cell-triggered inflammatory cell death
Nature Communications (2019)