Abstract
Receptors of the Notch family direct the differentiation of helper T cell subsets, but their influence on regulatory T cell (Treg cell) responses is obscure. We found here that lineage-specific deletion of components of the Notch pathway enhanced Treg cell–mediated suppression of type 1 helper T cell (TH1 cell) responses and protected against their TH1 skewing and apoptosis. In contrast, expression in Treg cells of a gain-of-function transgene encoding the Notch1 intracellular domain resulted in lymphoproliferation, exacerbated TH1 responses and autoimmunity. Cell-intrinsic canonical Notch signaling impaired Treg cell fitness and promoted the acquisition by Treg cells of a TH1 cell–like phenotype, whereas non-canonical Notch signaling dependent on the adaptor Rictor activated the kinase AKT–transcription factor Foxo1 axis and impaired the epigenetic stability of Foxp3. Our findings establish a critical role for Notch signaling in controlling peripheral Treg cell function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Yuan, J.S., Kousis, P.C., Suliman, S., Visan, I. & Guidos, C.J. Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 28, 343–365 (2010).
Radtke, F., Fasnacht, N. & Macdonald, H.R. Notch signaling in the immune system. Immunity 32, 14–27 (2010).
Perumalsamy, L.R., Marcel, N., Kulkarni, S., Radtke, F. & Sarin, A. Distinct spatial and molecular features of notch pathway assembly in regulatory T cells. Sci. Signal. 5, ra53 (2012).
Lee, K. et al. Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J. Exp. Med. 209, 713–728 (2012).
Elyaman, W. et al. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity 36, 623–634 (2012).
Osipo, C., Golde, T.E., Osborne, B.A. & Miele, L.A. Off the beaten pathway: the complex cross talk between Notch and NF-κB. Lab. Invest. 88, 11–17 (2008).
Poellinger, L. & Lendahl, U. Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Curr. Opin. Genet. Dev. 18, 449–454 (2008).
Amsen, D., Antov, A. & Flavell, R.A. The different faces of Notch in T-helper-cell differentiation. Nat. Rev. Immunol. 9, 116–124 (2009).
Cho, O.H. et al. Notch regulates cytolytic effector function in CD8+ T cells. J. Immunol. 182, 3380–3389 (2009).
Maekawa, Y. et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 9, 1140–1147 (2008).
Maekawa, Y. et al. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity 19, 549–559 (2003).
Minter, L.M. et al. Inhibitors of γ-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 6, 680–688 (2005).
Amsen, D. et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity 27, 89–99 (2007).
Tu, L. et al. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 202, 1037–1042 (2005).
Fang, T.C. et al. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity 27, 100–110 (2007).
Amsen, D. et al. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell 117, 515–526 (2004).
Keerthivasan, S. et al. Notch signaling regulates mouse and human Th17 differentiation. J. Immunol. 187, 692–701 (2011).
Mukherjee, S., Schaller, M.A., Neupane, R., Kunkel, S.L. & Lukacs, N.W. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J. Immunol. 182, 7381–7388 (2009).
Barbarulo, A. et al. Notch3 and canonical NF-κB signaling pathways cooperatively regulate Foxp3 transcription. J. Immunol. 186, 6199–6206 (2011).
Ou-Yang, H.F. et al. Notch signaling regulates the FOXP3 promoter through RBP-J- and Hes1-dependent mechanisms. Mol. Cell. Biochem. 320, 109–114 (2009).
Samon, J.B. et al. Notch1 and TGFβ1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood 112, 1813–1821 (2008).
Tran, I.T. et al. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J. Clin. Invest. 123, 1590–1604 (2013).
Sandy, A.R. et al. T cell-specific notch inhibition blocks graft-versus-host disease by inducing a hyporesponsive program in alloreactive CD4+ and CD8+ T cells. J. Immunol. 190, 5818–5828 (2013).
Roderick, J.E. et al. Therapeutic targeting of NOTCH signaling ameliorates immune-mediated bone marrow failure of aplastic anemia. J. Exp. Med. 210, 1311–1329 (2013).
Piggott, K. et al. Blocking the NOTCH pathway inhibits vascular inflammation in large-vessel vasculitis. Circulation 123, 309–318 (2011).
Shi, S. & Stanley, P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. USA 100, 5234–5239 (2003).
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002).
Yang, X. et al. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 269, 81–94 (2004).
Rubtsov, Y.P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Murtaugh, L.C., Stanger, B.Z., Kwan, K.M. & Melton, D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA 100, 14920–14925 (2003).
Hill, J.A. et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity 27, 786–800 (2007).
Marson, A. et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445, 931–935 (2007).
Sugimoto, N. et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int. Immunol. 18, 1197–1209 (2006).
Ouyang, W., Beckett, O., Flavell, R.A. & Li, M.O. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009).
Ouyang, W. et al. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature 491, 554–559 (2012).
Tejera, M.M., Kim, E.H., Sullivan, J.A., Plisch, E.H. & Suresh, M. FoxO1 controls effector-to-memory transition and maintenance of functional CD8 T cell memory. J. Immunol. 191, 187–199 (2013).
Zheng, Y. et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 463, 808–812 (2010).
Koch, M.A. et al. T-bet+ Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor β2. Immunity 37, 501–510 (2012).
Koch, M.A. et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 10, 595–602 (2009).
Bailis, W. et al. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity 39, 148–159 (2013).
Perumalsamy, L.R., Nagala, M., Banerjee, P. & Sarin, A. A hierarchical cascade activated by non-canonical Notch signaling and the mTOR-Rictor complex regulates neglect-induced death in mammalian cells. Cell Death Differ. 16, 879–889 (2009).
Huang, H. & Tindall, D.J. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim. Biophys. Acta 1813, 1961–1964 (2011).
Plas, D.R. & Thompson, C.B. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J. Biol. Chem. 278, 12361–12366 (2003).
Kerdiles, Y.M. et al. Foxo transcription factors control regulatory T cell development and function. Immunity 33, 890–904 (2010).
Floess, S. et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5, e38 (2007).
Ohkura, N. et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37, 785–799 (2012).
Oldenhove, G. et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009).
Zhou, X. et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10, 1000–1007 (2009).
Feuerer, M. et al. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc. Natl. Acad. Sci. USA 107, 5919–5924 (2010).
Li, Q.Z. et al. Protein array autoantibody profiles for insights into systemic lupus erythematosus and incomplete lupus syndromes. Clin. Exp. Immunol. 147, 60–70 (2007).
Schmitt, E.G. et al. IL-10 produced by induced regulatory T cells (iTregs) controls colitis and pathogenic ex-iTregs during immunotherapy. J. Immunol. 189, 5638–5648 (2012).
Noval Rivas, M. et al. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity 42, 512–523 (2015).
Acknowledgements
We thank C. Benoist for critical reading of the manuscript; P. Stanley (Albert Einstein College of Medicine) for Pofut1fl/fl mice; and T. Honjo (Kyoto University) for Rbpjfl/fl mice. Supported by the US National Institutes of Health (2R01AI065617, 2R01AI085090 and 1R56AI115699-01 to T.A.C.).
Author information
Authors and Affiliations
Contributions
L-.M.C. and T.A.C. designed the experiments and evaluated the data; L.-M.C., S.W., P.G. and E.S. performed experiments; L.-M.C. and E.S. analyzed data and prepared the figures; T.A.C. conceived of the project and directed the research; and L.-M.C. and T.A.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
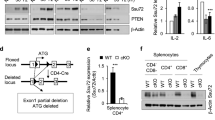
Supplementary Figure 1 Expression of Notch receptors on Tconv cells and Treg cells and the generation of mice with Treg cell–specific loss- and gain-of-function mutations in genes encoding components of the Notch pathway.
(a) Schematic representation of the derivation of mouse lines with TR cell-specific deletion of Pofut1, Rbpj, Notch1 and Rictor genes, obtained by crossing mice harboring a Foxp3EGFPCre bacterial artificial chromosome (BAC) with the respective LoxP flanked targeted gene strain. (b) EGFP+ and EGFP– CD4+ T cells were isolated by FACS sorting and levels of target gene transcripts were determined by real time PCR. Results were expressed as relative expression levels of the respective transcript compared to those of EGFP+ CD4+ T cells of Foxp3EGFPCre mice, which were given an average value of 1. (c) Tconv (CD4+EGFP–) and Treg (CD4+EGFP+) cells were FACS sorted from FoxP3EGFPcre reporter mice. Expression of Notch1-4 receptors was analyzed by real time PCR. Data are expressed as means +/– SEM (n= 4-5 independent samples). (ND = Not detected). (d) Left panel: Flow cytometric analysis on Notch1 expression on CD4+ Tconv and Treg cells from Foxp3EGFPCre mice. Right panel: bar graph representation of Notch1 mean fluorescence intensity in Tconv vs. Treg cells +/– SEM (n= 4 paired samples). (e) Schematic representation of the derivation of the mouse line with TR cell-specific expression of Notch1c. Foxp3EGFPCreRosa26N1c/N1c mice were obtained by crossing mice harboring Foxp3EGFPcre BAC with those carrying a knock-in allele at the Rosa26 locus encoding the cleaved intracellular domain of Notch1 (Notch1c) under control of a floxed stop signal (Rosa26N1c/N1c). (f) Flow cytometric analysis of Notch1c expression in Treg and Tconv cells (EGFP+ and EGFP–, respectively). (g) Heat map representation of transcript levels of Notch target genes, including Hes1, Hey1, Heyl and Dtx1, in Foxp3EGFPCre vs Foxp3EGFPCreRosa26N1c/N1c Treg cells revealed by gene expression array analysis. *p<0.05 by one way ANOVA with post test analysis or by unpaired two-tailed Student’s t-test.
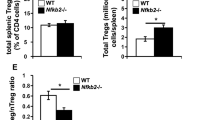
Supplementary Figure 2 Loss-of-function mutations in genes encoding components of the Notch signaling pathway promote the population expansion of Treg cells in the periphery in a cell-intrinsic manner.
(a) Flow cytometric analyses of CD4 and CD8 markers (upper panels) and Foxp3 on CD4 single positive (CD4SP) thymocytes (lower panels) in thymi of 8 weeks old Foxp3EGFPCre, Foxp3EGFPCrePofut1Δ/Δ, Foxp3EGFPCreRbpjΔ/Δ and Foxp3EGFPCreNotch1Δ/Δ mice. (b) Frequencies and numbers of thymic (Foxp3+CD4SP) Treg cells for each group. (c, d) Dose effect of TGF-β on induced Treg cell differentiation in vitro from naïve CD4+CD62L+CD44– T cells from Foxp3YFPCre, Foxp3YFPCreRbpjΔ/Δ and Foxp3YFPCreNotch1Δ/Δ. (e) Flow cytometric analyses of YFP and Foxp3 among CD4SP thymocytes (upper panels) or splenic CD4+ T cells in 8 weeks old heterozygous Foxp3YFPCre/WT and Foxp3YFPCre/WTRbpjΔ/Δ females. (f) Frequencies of YFP+ Treg cells within the thymus and the spleen for groups shown in (e). **p<0.01 by two way ANOVA with post test analysis.
Supplementary Figure 3 Notch1c impairs Treg cell fitness.
(a) Flow cytometric analyses of Foxp3 and EGFP expression in thymic and peripheral Treg cells (spleen) from Foxp3EGFPCreRosa26N1c/N1c mice. (b) Fractions of EGFP+ cells among Foxp3+ Treg cells shown in panel (a). (c) MFI of Notch1c in EGFP+ Treg cells from Foxp3EGFPcre and EGFP– and EGFP+ Treg cells from Foxp3EGFPCreRosa26N1c/N1c mice (d) Flow cytometric analyses of Foxp3 and fluorescent protein (EGFP or YFP) expression in peripheral Treg cells from Foxp3EGFPCreRosa26N1c/N1c and Foxp3YFPCreRosa26N1c/N1c mice. (e) Fractions of EGFP+ or YFP+ cells among Foxp3+ Treg cells shown in panel c. (f) Flow cytometric analyses of Foxp3 and YFP expression in peripheral Treg cells from Foxp3YFPCre/WTRosa26N1c/N1c heterozygotes females. (g) Fractions of YFP+ cells among Foxp3+ Treg cells shown in panel (f). The dashed blue line represents the 50% distribution expected from random X-inactivation. (h, i) Overlays and histograms of Foxp3 on YFP– and YFP+ Treg from Foxp3YFPCre/WTRosa26N1c/N1c from thymus, spleen and peripheral lymph nodes. ** p<0.01, *** p<0.001 by unpaired and paired Student’s two-tailed t test and one way ANOVA with post-test analysis.
Supplementary Figure 4 Accelerated development of severe lymphoproliferative and aggressive autoimmune multi-organ disease in Foxp3YFPCreRosa26N1c/N1c mice.
(a) Image of 8 weeks old Foxp3YFPCre and Foxp3YFPCreRosa26N1c/N1c male mice. (b) A representative picture of spleens and peripheral lymph nodes from 8 weeks old Foxp3YFPCre and Foxp3YFPCreRosa26N1c/N1c mice. (c) Survival curve of Foxp3YFPCre (n=6), Foxp3YFPCreRosa26N1c/N1c (n=9) and Foxp3YFPCreRosa26N1c/N1cRbpj∆/∆ (n=6) male mice. (d) Representative H&E stainings of ear, lung and colon of 8 weeks old Foxp3YFPCre and Foxp3YFPCreRosa26N1c/N1c mice. (e) Flow cytometric panels and (f) frequencies and numbers of splenic memory CD62LloCD44hi CD4 T cells from Foxp3YFPCre and Foxp3YFPCreRosa26N1c/N1c mice. (g) Flow cytometric panels and (h) frequencies and numbers of effector memory CD62LloCD44hi CD8 T cells. (i) Flow cytometric panels and (j) frequencies and numbers of IFN-γ and IL-17 producing CD4 T cells. (k) Flow cytometric panels and (l) frequencies and numbers of IFN-γ and IL-17 producing CD8 T cells. * p<0.05 and ** p<0.01 by unpaired Student’s two-tailed t test and log-rank test.
Supplementary Figure 5 Notch signaling impairs the Treg cell phenotype and the in vitro generation of Treg cells.
(a, b) Expression of Foxp3, CD25, CTLA4, Helios and Nrp1 markers were evaluated in Treg cells from 8 weeks old Foxp3YFPcre and Foxp3YFPCreRosa26N1c/N1c mice. Plain Gray represents expression of those markers in Tconv of Foxp3YFPCre mice. (c) Flow cytometric analyses of CD4 and CD8 markers (upper panels) and Foxp3 on CD4 single positive (CD4SP) thymocytes (lower panels) in thyme of 8 weeks old Foxp3EGFPCre and Foxp3EGFPCreRosa26N1c/N1c mice. (d) Frequencies and numbers of thymic (Foxp3+CD4SP) Treg cells for each group. (e) Heat map represent methylation status of individual CpG motifs within the TSDR of CNS2 in Foxp3. Individual CpG motifs are numbered with reference to the transcription initiation site of Foxp3 on thymic Treg cells of 8 weeks Foxp3EGFPCre and Foxp3EGFPCreRosa26N1c/N1c males. (f) Global methylation status of the TSDR of CNS2 in Foxp3. (g, h) Dose effect of TGF-β on induced Treg cell differentiation in vitro from naïve CD4+CD62LhiCD44lo T cells from Foxp3YFPCre and Foxp3YFPCreRosa26N1c/N1c. *p<0.05, **p<0.01 and ***p<0.001 by unpaired two-tailed Student’s t-test two way ANOVA.
Supplementary Figure 6 Notch1c mediates the cell-intrinsic destabilization of Treg cells.
(a) A representative picture of spleen and peripheral lymph nodes from 8 weeks old Foxp3YFPCre and Foxp3YFPCre/WTRosa26N1c/N1c and Foxp3YFPCreRosa26N1c/N1c females. (b) Dot plots represent total spleen cell number and frequencies of splenic naive (CD62LhiCD44lo), memory (CD62LloCD44hi) and IFN-γ+ CD4+ T cells for each group. Flow cytometric analyses of CD62L/CD44 (c) and IFN-γ/IL-17 (d) among CD4+ T cells for each group are shown. (e) Overlays of Treg cell canonical markers (Foxp3, CD25, CTLA4, OX40, Helios and Nrp1 on YFP– (Blue) and YFP+ (Red) from heterozygous Foxp3YFPCre/WTRosa26N1c/N1c females. Plain gray represent expression of those markers on CD4+ Tconv cells. (f) Dot plot represent MFI of Treg cell markers shown in (e) for YFP– and YFP+ Treg cells from heterozygous Foxp3YFPcre/WTRosa26N1c/N1c females. Representative overlays (g) and dot plots (h) of P-STAT4 on gated CD4+Foxp3-CXCR3+ (CD4+ Tconv), CD4+Foxp3+YFP–CXCR3+ (YFP– Treg), and CD4+Foxp3+YFP+CXCR3+ (YFP+ Treg) cells unstimulated (US) or upon IL-12 stimulation. * p<0.05, ** p<0.01 and *** p<0.001 by unpaired Student’s two-tailed t test, paired Student’s two-tailed t test and two way ANOVA with post-test analysis.
Supplementary Figure 7 RBPJ interferes with Foxp3 on Foxp3-binding sites and modulate Foxp3-mediated epigenetic signature on key Treg cell genes.
ChIP graphs represent quantitative PCR analysis of the ratio of enriched (a) Pde3b and (b) Ikzf2 Foxp3 binding sites immunoprecipitated with antibodies against Foxp3, RBPJ Histone H3 trimethylated at lysine 4 residu (H3K4me3) or dimethylated at lysine 27 residu (H3K27me2) (and respective control isotypes) to the input DNA on FACS sorted Treg cells isolated from Foxp3EGFPCre and Foxp3EGFPCreRosa26N1c/N1c mice (n=3 per group). ** p<0.01 and *** p<0.001 by unpaired two-tailed Student's t-test.
Supplementary Figure 8 Phenotypic analysis of Foxp3EGFPCreRosa26N1c/N1cRictorΔ/Δ Treg cells.
(a) Frequencies and numbers (b) of total CD4 T cells, memory CD4 T cells (CD62LloCD44hi), IFN-γ producing CD4 T cells and Treg cells in the spleen of 8 weeks old Foxp3EGFPCre, Foxp3EGFPCreRosa26N1c/N1c, Foxp3EGFPCreRosa26N1c/N1cRictorΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. (c) Frequencies and numbers (d) of total CD4 T cells, memory CD4 T cells (CD62LloCD44hi), IFN-γ producing CD4 T cells and Treg cells in the spleen of 8 weeks old Foxp3EGFPCre and Foxp3EGFPCreRictorΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. (e) Representative flow cytometric analyses of CD25, Nrp1, Helios, CTLA4 and Eos expression in splenic Treg cells of Foxp3EGFPCre, Foxp3EGFPCreRosa26N1c/N1c, Foxp3EGFPCreRosa26N1c/N1cRictorΔ/Δ and Foxp3EGFPCreRosa26N1c/N1cRbpjΔ/Δ mice. (f) Scatter plots of the MFI of individual markers in the respective mouse strains shown in (e). * p<0.05 **, p<0.01, *** p<0.001 by one way ANOVA with post test analysis.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 30133 kb)
Supplementary Table 1
Differentially expressed genes in Notch1c-overexpressing regulatory T cells (1.5X cut-off) (XLSX 76 kb)
Source data
Rights and permissions
About this article
Cite this article
Charbonnier, LM., Wang, S., Georgiev, P. et al. Control of peripheral tolerance by regulatory T cell–intrinsic Notch signaling. Nat Immunol 16, 1162–1173 (2015). https://doi.org/10.1038/ni.3288
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3288
This article is cited by
-
Non-canonical non-genomic morphogen signaling in anucleate platelets: a critical determinant of prothrombotic function in circulation
Cell Communication and Signaling (2024)
-
Therapy sculpts the complex interplay between cancer and the immune system during tumour evolution
Genome Medicine (2022)
-
Notch signaling pathway: architecture, disease, and therapeutics
Signal Transduction and Targeted Therapy (2022)
-
Isolation of nocobactin NAs as Notch signal inhibitors from Nocardia farcinica, a possibility of invasive evolution
The Journal of Antibiotics (2021)
-
A regulatory T cell Notch4–GDF15 axis licenses tissue inflammation in asthma
Nature Immunology (2020)