Abstract
CD4+ follicular helper T cells (TFH cells) are essential for germinal center (GC) responses and long-lived antibody responses. Here we report that naive CD4+ T cells deficient in the transcription factor Foxp1 'preferentially' differentiated into TFH cells, which resulted in substantially enhanced GC and antibody responses. We found that Foxp1 used both constitutive Foxp1A and Foxp1D induced by stimulation of the T cell antigen receptor (TCR) to inhibit the generation of TFH cells. Mechanistically, Foxp1 directly and negatively regulated interleukin 21 (IL-21); Foxp1 also dampened expression of the costimulatory molecule ICOS and its downstream signaling at early stages of T cell activation, which rendered Foxp1-deficient CD4+ T cells partially resistant to blockade of the ICOS ligand (ICOSL) during TFH cell development. Our findings demonstrate that Foxp1 is a critical negative regulator of TFH cell differentiation.
Similar content being viewed by others
Main
Help provided by CD4+ T cells to B cells is essential for the formation of germinal centers (GCs) and the generation of long-lived high-affinity antibodies. Follicular helper T cells (TFH cells) have been defined as a unique CD4+ T cell subset that provides such help to B cells1,2,3,4. TFH cells are characterized by the expression of molecules that facilitate functional interactions with B cells, including the chemokine receptor CXCR5, the cytokine interleukin 21 (IL-21) and the costimulatory molecules PD-1 and ICOS1,2,3,4,5,6,7,8. TFH cells also distinctively have high expression of the transcription factor Bcl-6, which has been demonstrated to be a central regulator of TFH cell differentiation9,10,11.
TFH cell differentiation has been proposed to be a multistage, multifactorial process4. Studies have shown that this differentiation involves interactions of CD4+ T cells with various types of antigen-presenting cells, such as dendritic cells (DCs) and B cells8,12,13,14,15. The presentation of antigen by DCs is necessary and sufficient to initiate the TFH cell–differentiation program consisting of early induction of the expression of CXCR5, Bcl-6 and ICOS14,15,16. The interaction of ICOS with its ligand ICOSL is critical in 'instructing' TFH cell differentiation; in the absence of ICOS or in the presence of blocking antibodies to ICOSL, TFH cell differentiation is substantially impaired8,14. After the DC priming stage, further TFH cell differentiation involves a B cell–dependent stage9,14,15,16,17 in which signaling via ICOS is required for both the maintenance of Bcl-6 expression in TFH cells and the follicular relocation of TFH cells into GCs14,16,18. In the absence of B cells, DC-initiated TFH cell responses are aborted14,15.
In addition to antigen-presenting cells and costimulation via ICOS, the cytokine milieu has important roles in TFH cell differentiation7,8,19,20,21,22,23. IL-6 and IL-21 (which engage the pathways of the signal transducers STAT1 and STAT3) and IL-2 (which engages the STAT5 pathway) have been shown to favor TFH cell differentiation and limit it, respectively7,8,19,20,21. IL-21 also acts directly on B cells at various stages of GC B cell responses24,25,26. At the transcriptional level, Bcl-6 and its antagonist Blimp-1 have central roles in TFH cell differentiation9. Several other transcription factors (Batf, Irf4, c-Maf and Ascl2) are also important for TFH cell development27,28,29,30,31. Despite all these findings, the molecular mechanisms that underlie TFH cell differentiation, particularly initial TFH cell development, have remained unclear.
The forkhead box ('Fox') proteins constitute a large family of transcription factors with diverse functions32,33. Foxp1, a member of the 'Foxp' subfamily, is expressed in many tissues and has four isoforms (Foxp1A, Foxp1B, Foxp1C and Foxp1D)34. In cells of the T lineage, Foxp1 has important roles in both the generation of quiescent naive T cells and the maintenance of naive T cell quiescence in the periphery35,36.
Here we report that in a T cell–dependent immune response, Foxp1 was a rate-limiting and critical negative regulator of TFH cell differentiation. We found that in addition to using its constitutive Foxp1A isoform, Foxp1 also used a Foxp1D isoform induced by stimulation via the T cell antigen receptor (TCR) to efficiently block initial TFH cell development and that the negative regulation of TFH cell differentiation by Foxp1A and Foxp1D was dose dependent. Mechanistically, we found that Foxp1 directly and negatively regulated IL-21 and that Foxp1 dampened the expression of ICOS and its downstream signaling, which resulted in partial resistance of Foxp1-deficient CD4+ T cells to blockade of ICOSL during TFH cell development. The negative regulation of TFH cell differentiation by Foxp1 also showed profound dominance, such that even in the absence of B cells, Foxp1-deficient CD4+ T cells differentiated into TFH cells at high frequencies with sustained Bcl-6 expression. Our findings demonstrate that the two Foxp1 isoforms provide a 'double-check' mechanism for fundamental regulation of TFH cell differentiation and humoral responses.
Results
TCR stimulation transiently induces Foxp1D expression
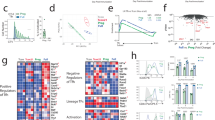
To study how Foxp1 regulates the responses of CD4+ T cells to challenge with antigen, we first examined Foxp1 expression patterns during the activation of CD4+ T cells. We found that in wild-type naive CD4+ T cells, upon stimulation in vitro with antibody to the invariant signaling protein CD3 (anti-CD3) and antibody to the coreceptor CD28 (anti-CD28), expression of constitutive full-length Foxp1A was constant; conversely, among the other three shorter isoforms, expression of only Foxp1D was induced (Fig. 1a). Consistent with those immunoblot analysis results, intracellular staining revealed increased total Foxp1 protein following activation (Fig. 1b), reflective of the induction of Foxp1D expression. The TCR-induced expression of Foxp1D decreased when the TCR stimulation was withdrawn (Fig. 1c), which suggested that sustained Foxp1D expression was dependent on the duration of TCR stimulation. Nevertheless, once T cells were activated (as indicated by expression of the activation marker CD44), both low and high doses of TCR stimulation induced similar expression of Foxp1D protein (Supplementary Fig. 1a).
(a) Immunoblot analysis of Foxp1 in wild-type (WT) and Foxp1-deficient (cKO) CD4+ T cells left inactivated (0) or activated for 2–5 d in vitro; β-actin serves as a loading control throughout. (b) Intracellular staining of Foxp1 in wild-type and Foxp1-deficient CD4+ T cells left inactivated (naive) or activated for 2 d in vitro and labeled with the fluorescent stain CellTrace. (c) Immunoblot analysis of Foxp1 in wild-type CD4+ T cells stimulated for 1 d or 2 d with plate-bound anti-CD3 (sustain (2(s))) or stimulated for 1 d with plate-bound anti-CD3, followed by transfer of cells to a new well for another day without further stimulation with anti-CD3 (detach (2(d))). (d) Intracellular staining of Foxp1 in naive host CD4+ T cells (Endo) and donor OT-II T cells (OT-II) obtained from Ly5.1+ C57BL/6 mice given CellTrace-labeled wild-type OT-II T cells, assessed at day 2 or 3 (above plots) after immunization of recipients with NP-OVA in alum. (e) Immunoblot analysis of Foxp1 in donor OT-II T cells obtained from Ly5.1+ C57BL/6 mice given wild-type OT-II T cells, assessed before (0) or 3 d after immunization with NP-OVA in alum. Data represent at least two independent experiments.
To demonstrate that the induction of Foxp1D expression in activated CD4+ T cells also occurred in vivo, we transferred naive OT-II T cells (which have transgenic expression of an ovalbumin (OVA)-specific TCR) into Ly5.1+ C57BL/6 recipient mice, followed by immunization of the recipients with OVA conjugated to 4-hydroxy-3-nitrophenylacetyl (NP-OVA) in alum. At 2 d after immunization, intracellular staining showed that the total amount of Foxp1 in proliferating donor OT-II T cells had increased, and by day 3, it started to decrease (Fig. 1d). Immunoblot analysis revealed that Foxp1D expression was indeed induced in donor OT-II T cells at day 3, with almost no change in Foxp1A expression (Fig. 1e). By days 4–5, Foxp1 in donor OT-II T cells decreased to amounts equivalent to those of naive T cells (Supplementary Fig. 1b). Thus, during the initial days of an in vivo immune response, Foxp1D expression was transiently induced in CD4+ T cells by antigen stimulation.
Foxp1 negatively regulates TFH cell differentiation
We crossed mice that undergo conditional deletion of loxP-flanked Foxp1 alleles (Foxp1f/f) mediated by Cre recombinase expressed from the gene encoding a tamoxifen-sensitive estrogen receptor variant (Cre-ERT2) and express yellow fluorescent protein (YFP) from the ubiquitously expressed Rosa26 locus (Foxp1f/fCre-ERT2+RosaYFP mice)36 with OT-II mice to generate OT-II Foxp1f/fCre-ERT2+RosaYFP mice. We sorted CD44loCD62LhiCD25−YFP+ naive Foxp1-deficient OT-II (OT-II Foxp1-cKO) T cells or naive Foxp1-sufficient (Foxp1–wild-type) OT-II (OT-II Foxp1-WT) T cells from tamoxifen-treated OT-II Foxp1f/fCre-ERT2+RosaYFP mice or from control OT-II Foxp1f/fRosaYFP mice (or OT-II Foxp1+/+Cre-ERT2+RosaYFP mice), respectively. We then transferred the sorted cells into SMARTA mice (which have transgenic expression of a TCR specific for the lymphocytic choriomeningitis virus epitope of glycoprotein amino acids 66–77), followed by immunization of the recipient mice with NP-OVA in alum. We used SMARTA mice as recipients to reduce the competition between transferred donor OT-II T cells and host T cells in response to challenge with NP-OVA13. In the subsequent 2 weeks after immunization, OT-II Foxp1-cKO T cells and OT-II Foxp1-WT T cells responded with similar kinetics and magnitudes of population expansion and contraction (Fig. 2a). However, the OT-II Foxp1-cKO T cell population contained a much higher frequency and number of CXCR5hiPD1hi TFH cells, with a correspondingly lower frequency and number of non-TFH cells, than that of the OT-II Foxp1-WT T cell population (Fig. 2a,b). On the basis of staining for the marker GL7 and CXCR5 (ref. 37), OT-II Foxp1-cKO T cell population also included a higher frequency of GC TFH cells (Fig. 2c). OT-II Foxp1-cKO and OT-II Foxp1-WT TFH cells were phenotypically indistinguishable (CD44hiCD62LloICOShiBTLAhiCD200hi; Fig. 2d) and expressed similar amounts of Bcl-6 protein (Fig. 2e). Experiments with Ly5.1+ C57BL/6 mice as recipients resulted in a similarly increased frequency of TFH cells among Foxp1-deficient OT-II T cells but a lower magnitude of response than that observed with SMARTA mice (Supplementary Fig. 2a–c and data not shown). These results suggested that deletion of Foxp1 in naive CD4+ T cells led to the 'preferential' development of TFH cells and GC TFH cells at the expense of non-TFH cells. Thus, Foxp1 had a negative regulatory role in TFH cell development.
(a) Quantification of total splenic OT-II T cells, TFH cells and non-TFH cells obtained from Ly5.1+ SMARTA mice given transfer of purified naive OT-II Foxp1-WT or OT-II Foxp1-cKO T cells (key), followed by immunization with NP-OVA in alum and analysis 3–14 d later. (b,c) Flow cytometry of donor OT-II T cells obtained from the mesenteric lymph nodes (mLN) and spleen (Spl) of recipient mice at 5 d (b) or 7 d (c) after immunization as in a. Numbers adjacent to outlined areas indicate percent PD-1hiCXCR5hi TFH cells (b) or GL7hiCXCR5hi GC TFH cells (c). (d) Phenotype of OT-II TFH cells at day 5 after immunization as in a, assessed by staining for various markers (horizontal axes). (e) Intracellular staining of Bcl-6 in OT-II T cells at day 7 after immunization as in a (left); numbers adjacent to outlined areas indicate percent Bcl-6+CXCR5hi TFH cells. Right, overlay of histograms of TFH cells. *P < 0.05 and **P < 0.01 (two-tailed Student's t-test). Data represent at least three independent experiments (error bars (a), s.d. of four (day 3, 5 or 7) or three (day 14) mice per group).
Studies have shown that Foxp3+ follicular regulatory T cells have an important role in suppressing TFH cell development38,39. We stained cells for Foxp3 and found that neither OT-II Foxp1-WT T cells nor OT-II Foxp1-cKO T cells generated any Foxp3+ OT-II follicular regulatory T cells in SMARTA recipient mice (Supplementary Fig. 3a). There was also no difference between the two groups of SMARTA recipient mice in their regulatory T cell populations (Supplementary Fig. 3a). Finally, we transferred OT-II Foxp1-WT T cells and OT-II Foxp1-cKO T cells together into the same recipient mouse and found that the Foxp1-deficient OT-II T cell population still contained a substantially higher frequency of TFH cells (Supplementary Fig. 3b), which suggested that the effect of Foxp1 on TFH differentiation was cell intrinsic. As for the role of Foxp1 in the differentiation of other subsets of CD4+ T cells, we found that while the loss of Foxp1 did not seem to alter differentiation of the TH1 or TH17 subset of helper T cells in vitro, it might have slightly favored the differentiation of TH2 cells (Supplementary Fig. 4).
Foxp1-deficient TFH cells lead to enhanced GC responses
Consistent with the enhanced TFH cell response noted above, both the frequency and number of GC B cells in the recipient mice that received OT-II Foxp1-cKO T cells were higher than those in mice that received OT-II Foxp1-WT T cells (Fig. 3a). Histological analysis of spleen sections 6 d after immunization showed that the mice that received OT-II Foxp1-cKO T cells had an increased size and frequency of GCs (Fig. 3b). By day 6 after immunization with antigen, more OT-II Foxp1-cKO T cells than OT-II Foxp1-WT T cells had already localized to the follicles and GCs (Fig. 3b), which suggested that Foxp1-deficient TFH cells acted anatomically and functionally like TFH cells.
(a) Flow cytometry of donor OT-II T cells from the mesenteric lymph nodes and spleens of Ly5.1+ SMARTA recipient mice given transfer of purified naive OT-II Foxp1-WT or OT-II Foxp1-cKO T cells, followed by immunization of the recipients with NP-OVA in alum and analysis 7 d later. Numbers adjacent to outlined areas (left) indicate percent GC B cells (Fas+PNA+) among total B cells (B220+). Right, total GC B cells. (b) Confocal microscopy of B cell follicles (IgD+), GCs (AID+) and the localization of donor (Ly5.2+) OT-II Foxp1-WT or OT-II Foxp1-cKO T cells in Ly5.1+ SMARTA recipient mice at 6 d after immunization as in a. Scale bars, 500 μm (top) or 100 μm (bottom). (c) Quantification of antibody-secreting cells (ASC) producing NP25-specific IgM, IgG or IgG1 in the spleens of recipient mice 7 d after immunization as in a. (d) Kinetics of the production of titers of low-affinity (NP25) and high-affinity (NP4) NP-specific IgG in the serum of the recipient mice in a. *P < 0.05 and **P < 0.01 (two-tailed Student's t-test). Data represent at least two independent experiments (error bars, s.d. of four mice per group (a), three mice per group (c) or two (day 5) or three (day 7, 8, 14 or 21) mice per group (d)).
By day 7 after immunization, although the number of antibody-secreting cells that produced NP-specific immunoglobulin M (IgM) was similar in both groups of recipient mice (Fig. 3c), the number of antibody-secreting cells producing NP-specific total IgG and IgG1 in mice that received OT-II Foxp1-cKO T cells was almost tenfold greater than that in their counterparts that received OT-II Foxp1-WT T cells (Fig. 3c). NP-specific IgG antibodies of both low affinity (antibodies to a NP25-BSA (NP conjugated to bovine serum albumin (BSA) at a molecular ratio of 25:1)) and high affinity (antibodies to NP4-BSA (NP conjugated to BSA at a molecular ratio of 4:1)) were also produced at much higher titers in recipients of OT-II Foxp1-cKO T cells (Fig. 3d). Thus, the enhanced TFH cell responses of Foxp1-deficient CD4+ T cells correlated with greatly increased GC responses and the production of antibodies of both low affinity and high affinity.
TCR-induced Foxp1D blocks initial TFH differentiation
Given the implications of the differences in isoform expression following stimulation of the TCR (Fig. 1a,b,d,e), we investigated the individual roles of Foxp1A and Foxp1D in TFH cell differentiation. We generated mice with conditional transgenic expression of the specific isoforms Foxp1A and Foxp1D through the use of a Rosa26 locus–knock-in approach ('Foxp1aTg/Tg mice' and 'Foxp1dTg/Tg mice', respectively; Supplementary Fig. 5a,b). We crossed those mice with mice that express Cre recombinase from the T cell–specific Cd4 promoter (Cd4-Cre) to generate Foxp1aTg/TgCd4-Cre mice and Foxp1dTg/TgCd4-Cre mice, respectively. Naive T cells from both mouse lines developed in normal numbers with a normal phenotype and expressed green fluorescent protein (GFP) as a marker (Supplementary Fig. 5c,d). In addition to using the model antigen OVA, we also extended the studies and used a model of infection with influenza virus. We infected Foxp1aTg/TgCd4-Cre and Foxp1dTg/TgCd4-Cre mice with influenza virus strain A/Puerto Rico/8/34 (PR8) and found that both transgenes potently suppressed the generation of TFH cells and subsequent GC B cell responses (Supplementary Fig. 6a–d). We also observed suppression of TFH cell development by the transgene encoding Foxp1D in naive OT-II T cells from OT-II Foxp1dTg/TgCd4-Cre mice in the OVA model (Supplementary Fig. 7a). The suppression of TFH cell development by Foxp1D was specific rather than being a general suppression of CD4+ T cell responses, as the overall magnitude of the CD4+ T cell responses was similar for wild-type OT-II T cells and OT-II T cells with transgenic expression of Foxp1D when they were transferred together into the same Ly5.1+ C57BL/6 recipient mice, followed by immunization of the recipients with NP-OVA in alum (Supplementary Fig. 7b). Together our results suggested that Foxp1, including both constitutively expressed Foxp1A and TCR-induced Foxp1D, had a negative role in TFH cell development.
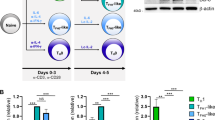
In the OT-II OVA model, TFH cells began to develop around day 3 (ref. 40) (Fig. 2a), the time at which Foxp1D expression began to decrease from its peak at day 2 (Fig. 1d). To achieve Foxp1D-specific deletion and address whether Foxp1D induced by stimulation of the TCR has a role in blocking initial TFH cell development, we generated OT-II Foxp1aTg/+Foxp1f/fCd4-Cre (OT-II Foxp1D-KO) mice. This resulted in deletion of all endogenous Foxp1 with maintenance of T cell–specific expression of the transgene encoding Foxp1A (Fig. 4a,b). In recipient mice given OT-II Foxp1-WT T cells or OT-II Foxp1D-KO T cells, at 3 d after immunization with NP-OVA in alum, only a small proportion of OT-II Foxp1-WT T cells differentiated into CXCR5hiPD1hi TFH cells (Fig. 4c), whereas almost 40% of the donor OT-II Foxp1D-KO T cells developed into TFH cells (Fig. 4c). This occurred even though the abundance of transgenically expressed Foxp1A protein in OT-II Foxp1D-KO T cells was higher than the abundance of endogenous Foxp1A in OT-II Foxp1-WT T cells (Fig. 4a). By day 7, the frequency of OT-II Foxp1D-KO TFH cells was still higher than that of OT-II Foxp1-WT control cells (Fig. 4c). These results demonstrated that during the early stages of a T cell–dependent response, Foxp1D induced by antigen stimulation was particularly critical in blocking initial TFH cell differentiation.
(a,b) Immunoblot analysis (a) and intracellular staining (b) of Foxp1 in donor OT-II T cells from Ly5.1+ SMARTA mice given transfer of purified naive OT-II Foxp1-WT (WT), OT-II Foxp1-Het (Het), OT-II Foxp1D-KO (1D-KO) or OT-II Foxp1-cKO (cKO) T cells, followed by immunization with NP-OVA in alum and analysis 3 d later. Naive (b), naive CD4+ (endogenous) T cells from recipient mice. (c,d) TFH differentiation of donor OT-II T cells in the spleens of the recipient mice in a,b at day 3 or 7 (c) or day 3 or days 7–8 (d) after immunization. Numbers adjacent to outlined areas (left) indicate percent PD-1hiCXCR5hi TFH cells. *P < 0.05 and **P < 0.01 (two-tailed Student's t-test). Data represent at least two independent experiments (error bars (c,d), s.d. of four mice per group).
Sum of Foxp1 proteins serves as a rate-limiting factor
Knowing that both Foxp1A and Foxp1D inhibited TFH cell differentiation (Supplementary Fig. 6a,c), we next sought to determine whether the 'dose' of Foxp1 protein was critical for TFH cell development. The amount of Foxp1A in naive T cells from mice with heterozygous Foxp1 expression is lower than that in wild-type T cells36. Here we found that both Foxp1A and Foxp1D had lower expression in activated CD4+ T cells from such heterozygous mice than in their wild-type counterparts (Fig. 4a,b). We transferred naive OT-II T cells from tamoxifen-treated OT-II Foxp1f/fRosaYFP (OT-II Foxp1-WT) mice, OT-II Foxp1f/+Cre-ERT2+RosaYFP (OT-II Foxp1-Het) mice or OT-II Foxp1f/fCre-ERT2+RosaYFP (OT-II Foxp1-cKO) mice into SMARTA mice and immunized the recipients with NP-OVA in alum. At either day 3 or days 7–8 after immunization, the frequency of OT-II Foxp1-Het TFH cells was intermediate between that of OT-II Foxp1-WT TFH cells and that of OT-II Foxp1-cKO TFH cells (Fig. 4d). These results demonstrated dose-dependent regulation of TFH cell differentiation by Foxp1 and suggested that the sum of total Foxp1 served as a rate-limiting factor for TFH cell development.
Foxp1-deficient TFH cells are true TFH cells
To initially address the mechanism underlying the substantial effect of Foxp1 on TFH cell development, we analyzed global gene expression in TFH cells generated in vivo. We identified a group of 118 highly TFH cell–specific genes by combining gene-expression data obtained with OT-II Foxp1-WT TFH cells and three publically available data sets of TFH cell expression (GEO accession codes GSE24574, GSE16697 and GSE40068; Fig. 5a and Supplementary Table 1). We found high concordance in the expression profiles of wild-type and Foxp1-deficient CXCR5hiPD1hi TFH cells (Pearson r2 = 0.90), and most of the 118 TFH cell–specific genes and genes encoding some additional markers had similar expression in the two groups of TFH cells (Fig. 5a and Supplementary Table 1). Thus, phenotypically, anatomically and functionally, Foxp1-deficient TFH cells were very similar to wild-type TFH cells.
(a) Microarray analysis of the expression of 118 genes encoding TFH cell markers (blue dots; left) and genes encoding additional markers in the literature (red dots; left) in OT-II Foxp1-WT and OT-II Foxp1-cKO TFH cells at day 5 ex vivo. Right, heat map of the expression, in OT-II Foxp1-WT and OT-II Foxp1-cKO TFH cells (TFH; replicates 1 and 2) and non-TFH cells (n; replicate 1), of the top 20 genes (with the greatest difference in expression in wild-type TFH cells relative to their expression in non-TFH cells) encoding TFH cell markers. (b) Intracellular staining of IL-21 in recipient CD4+ T cells (far left) and donor OT-II T cells from the spleen of Ly5.1+ SMARTA mice given transfer of purified naive OT-II Foxp1-WT or OT-II Foxp1-cKO T cells, followed by immunization with NP-OVA in alum and analysis 7 d later. Numbers adjacent to outlined areas (left) indicate percent IL-21+ cells. FSC, forward scatter. (c,d) Intracellular staining of interferon-γ (IFN-γ) and IL-21 (c) and real time-PCR analysis of Il21 mRNA (d) in naive Foxp1-WT or Foxp1-cKO CD4+ T cells stimulated in vitro for 2 d with anti-CD3 and anti-CD28 under TH1-polarizing culture conditions, followed by population expansion for additional 2 d in medium and analysis on day 4. Numbers in quadrants (c) indicate percent cells in each; mRNA expression (d) was normalized to that of mRNA encoding the ribosomal protein L32 (Rpl32 mRNA) and is presented relative to that of Foxp1-WT cells. (e) Predicted forkhead-binding site (underlined) in the Il21 promoter (left), and chromatin-immunoprecipitation analysis of the binding of Foxp1 to a control region (Ctrl) or the promoter region (Prom) of Il21, presented as binding in Foxp1-WT T cells relative to binding in Foxp1-cKO T cells (right). *P < 0.05 and **P < 0.01 (two-tailed Student's t-test). Data represent two (a,e) or at least three (b–d) independent experiments (error bars, s.d. of four (b) or three (d) mice per group).
In addition to assessing the 118 TFH cell–specific genes, we also assessed mRNA encoding a group of key molecules known to be critical for TFH cell differentiation. We compared the abundance of these mRNAs in wild-type and Foxp1-deficient CD4+ T cells activated in vitro under conditions that promote the differentiation of TFH cell–like cells4. Consistent with the results obtained for TFH cells ex vivo (Fig. 5a), all the genes analyzed by reverse transcription followed by quantitative PCR had similar expression in the two groups of TFH cell–like cells in vitro (Supplementary Fig. 8a).
Foxp1 directly and negatively regulates IL-21
A few of the 118 TFH cell–specific genes had higher expression in Foxp1-deficient TFH cells than in wild-type TFH cells, and one of these was the gene encoding IL-21 (Fig. 5a and Supplementary Table 1). Indeed, we did find that a greater proportion of ex vivo OT-II Foxp1-cKO T cells than OT-II Foxp1-WT T cells produced IL-21 (Fig. 5b), which suggested that Foxp1 may have negatively regulated IL-21. The TH1 cell–inducing cytokine IL-12 has been shown to induce IL-21 production22,23. Thus, we examined IL-21 production under TH1-polarizing culture conditions in vitro. We found that a higher frequency of Foxp1-deficient TH1 cells than wild-type cells produced IL-21 at the level of both protein and mRNA (Fig. 5c,d). Notably, under the same TH1-polarizing culture conditions, the transgene encoding Foxp1D specifically suppressed the production of IL-21 but not the production of interferon-γ (Supplementary Fig. 8b), which suggested that IL-21 may have been directly regulated by Foxp1. Bioinformatics analysis identified one highly conserved forkhead-binding consensus site in the promoter region of the locus encoding IL-21 across species (Fig. 5e). Chromatin-immunoprecipitation analysis of Foxp1 in wild-type TH1 cells showed that Foxp1 bound specifically to the Il21 promoter region (Fig. 5e). Together these in vivo and in vitro results suggested that Foxp1 negatively and directly regulated IL-21.
Partial resistance to blockade of ICOSL
Foxp1 did not seem to regulate ICOS expression directly (Fig. 5a and Supplementary Fig. 8a). However, by examining the OT-II T cells at the early time points after T cell activation, we found that ICOS expression was higher in Foxp1-cKO T cells than in Foxp1-WT T cells both in vitro and in vivo (Fig. 6a,b and Supplementary Table 2). This suggested that the Foxp1 pathway indirectly dampened the initial cell-surface abundance of ICOS during T cell activation.
(a) Staining of ICOS in purified naive Foxp1-WT and Foxp1-cKO CD4+ T cells mixed at ratio of 1:1 and activated for 1–3 d by stimulation in vitro with anti-CD3 and anti-CD28. (b,c) Staining of ICOS (b) and intracellular staining of Foxo1 (c) in naive donor OT-II Foxp1-WT and OT-II Foxp1-cKO T cells mixed at ratio of 1:1 and transferred together into Ly5.1+ C57BL/6 mice, followed by immunization of the recipients with NP-OVA in alum, assessed at days 2–4 (b) or day 3 (c) after immunization. (d) Flow cytometry of donor OT-II T cells (left) from Ly5.1+ SMARTA mice given transfer of purified naive OT-II Foxp1-WT or OT-II Foxp1-cKO T cells, followed by immunization of recipient mice with NP-OVA in alum and treatment with PBS or anti-ICOSL, and analysis at day 4 after immunization. Numbers adjacent to outlined areas (left) indicate percent PD-1hiCXCR5hi TFH cells. Right, extent of the blockade of the TFH differentiation of donor OT-II T cells after treatment with anti-ICOSL. (e) Flow cytometry of donor OT-II T cells obtained as in d. Numbers adjacent to outlined areas indicate percent Bcl-6+CXCR5hi TFH cells at day 4 after immunization. *P < 0.01 (two-tailed Student's t-test). Data represent two (a,e) or at least three (b–d) independent experiments (error bars (d), s.d. of five mice per group).
It has been reported that ICOS-mediated signaling via phosphatidylinositol-3-OH kinase is critical for the generation of TFH cells41. Activation of signaling via phosphatidylinositol-3-OH kinase and the kinase Akt induces the phosphorylation of Foxo1 and leads to the degradation of Foxo1 protein33. Therefore, we assessed the activation of Foxo1 in OT-II T cells in recipient mice given transfer of OT-II Foxp1-WT or OT-II Foxp1-cKO T cells, followed by immunization of the recipients with NP-OVA in alum and analysis 3 d later. In donor OT-II T cells at day 3 ex vivo, we found that the amount of total Foxo1 was slightly but significantly lower in Foxp1-deficient cells than in wild-type control cells (Fig. 6c and Supplementary Table 2), which suggested that, consistent with the enhanced induction of ICOS, downstream signaling via ICOS in Foxp1-deficient OT-II T cells was also more activated. The negative regulation of ICOS expression and signaling via ICOS by Foxp1 led us to hypothesize that the TFH differentiation of Foxp1-deficient T cells would be resistant to blockade of ICOSL. In recipient mice given transfer of OT-II Foxp1-WT or OT-II Foxp1-cKO T cells and treated with antibodies to ICOSL, we found that the antibodies blocked 70–80% of the TFH differentiation of OT-II Foxp1-WT T cells; however, such antibodies blocked only about 30% of the TFH differentiation of OT-II Foxp1-cKO T cells (Fig. 6d), and OT-II Foxp1-cKO TFH cells still expressed Bcl-6 (Fig. 6e). Collectively, these results suggested that Foxp1-deficient CD4+ T cells were partially resistant to the blockade of ICOSL during TFH differentiation.
Profound effect of Foxp1 on TFH cell differentiation
After the initial DC priming stage and during the T cell–B cell interaction stage, ICOS signaling has been shown to have critical roles in maintaining TFH cell differentiation8,14. The partial resistance of the TFH differentiation of Foxp1-deficient CD4+ T cells to the blockade of ICOSL led us to investigate whether Foxp1-deficient CD4+ T cells would still differentiate into TFH cells in the absence of B cells. We found that, as reported14, in B cell–deficient μMT recipient mice given OT-II Foxp1-WT T cells or OT-II Foxp1-cKO T cells and then challenged with NP-OVA in alum, the TFH development of OT-II Foxp1-WT T cells was aborted (Fig. 7a). Strikingly, compared with OT-II Foxp1-WT control cells, OT-II Foxp1-cKO T cells not only elicited enhanced TFH cell responses in μMT recipient mice (Fig. 7b) but also exhibited TFH cell frequencies even higher than those of OT-II Foxp1-WT T cells in B cell–intact recipient mice (Fig. 7a). The finding that the total number of OT-II Foxp1-cKO TFH cells was lower in μMT recipient mice than in B cell–intact recipient mice (Figs. 2a and 7b) might have resulted from the much lower number of total donor OT-II T cells (Foxp1-cKO as well as Foxp1-WT) at the early stage (day 3) of the response after transfer (Supplementary Fig. 9). The decrease in the OT-II Foxp1-cKO TFH cell response seemed to be sharper in μMT recipient mice than in B cell–intact recipient mice (Figs. 2a and 7b), which suggested that B cells may be important for the survival of TFH cells. Nevertheless, given that the TFH cell response of OT-II Foxp1-WT T cells in μMT recipient mice was aborted (Fig. 7a,b), the induced OT-II Foxp1-cKO TFH cell response in μMT recipient mice was notable. In μMT recipient mice, OT-II Foxp1-cKO TFH cells exhibited a conventional TFH cell phenotype, with 10–20% even able to convert into GL7hiCXCR5hi GC TFH cells (Supplementary Fig. 10a,b). The frequency of IL-21-producing cells was also higher among OT-II Foxp1-cKO T cells than among OT-II Foxp1-WT T cells (Supplementary Fig. 10c). At later time points, when the responses were diminishing, OT-II Foxp1-cKO TFH cells continued to express Bcl-6 in the absence of B cells (Fig. 7c). Thus, even in the absence of B cells, Foxp1-deficient T cells generated substantial TFH cell responses.
(a,b) Frequency (a) and number (b) of donor OT-II TFH cells obtained from the spleen of Ly5.1+ C57BL/6 (B6) or μMT mice given transfer of purified naive OT-II Foxp1-WT T cells (WT→) or OT-II Foxp1-cKO T cells (cKO→), followed by immunization with NP-OVA in alum and analysis at day 4 (a) or days 3–7 (b) after immunization. (c) Intracellular staining of Bcl-6 in donor OT-II T cells obtained from host mice on day 4 or day 7 after immunization as in a. (d) Flow cytometry of retrovirus-infected donor OT-II T cells from the mesenteric lymph nodes of Ly5.1+ C57BL/6 recipient mice given transfer of OT-II Foxp1-WT or OT-II Foxp1-cKO T cells infected with control retrovirus (RV-Ctrl) or retrovirus expressing Bcl-6 (RV–Bcl-6) or Foxp1A (RV-Foxp1A), followed by challenge of the recipient mice with NP-OVA in alum 1 or 2 d after cell transfer ('rest'; left margin) and analysis 5 d after immunization. (e) Flow cytometry of retrovirus-infected donor (SMARTA) T cells from the spleens of Ly5.1+ C57BL/6 recipient mice given transfer of CD4+ T cells from SMARTA Foxp1f/f mice infected with control retrovirus (RV-Ctrl) or retrovirus expressing Cre (RV-Cre) or Bcl-6 (RV–Bcl-6), followed by immunization of the recipient mice 2 d later with gp61-KLH in alum and analysis 5 d after immunization. Right, frequency of TFH cells. Numbers adjacent to outlined areas indicate percent PD-1+CXCR5+ (TFH) cells (a,d,e) or Bcl-6+CXCR5+ cells (c). *P < 0.05 and **P < 0.01 (two-tailed Student's t-test). Data represent two independent experiments (error bars (b,e), s.d. of two (day 3) or three (days 4, 5 and 7) mice per group (b) or five mice per group (e)).
The substantial effect of Foxp1 on TFH cell differentiation in the absence of B cells led us to compare the TFH development of Foxp1-deficient T cells with overexpression of Bcl-6. As reported for cells in vivo9,18,29, overexpression of Bcl-6 in OT-II Foxp1-WT T cells enhanced their TFH development (Fig. 7d and Supplementary Fig. 11a–c). However, unexpectedly, the frequency of OT-II TFH cells generated in the absence of Foxp1 was much higher than that of OT-II Foxp1-WT T cells that overexpressed Bcl-6 (Fig. 7d and Supplementary Fig. 11c). We obtained similar results with either Ly5.1+ C57BL/6 mice or SMARTA mice as the recipients (data not shown). Retroviral overexpression of Foxp1A (or Foxp1D) 'rescued' the abnormally enhanced TFH differentiation of OT-II Foxp1-cKO T cells (Fig. 7d, Supplementary Fig. 11c and data not shown).
In addition to assessing this in OT-II T cells, we also investigated the function of Foxp1 in the TFH differentiation of SMARTA T cells. We introduced Cre expression into SMARTA Foxp1f/f T cells in vitro by retroviral infection after T cells were activated and then transferred the infected T cells into the Ly5.1+ C57BL/6 recipient mice. We found that the Foxp1-deficient SMARTA T cell population also contained a higher frequency of TFH cells after immunization of recipient mice with the lymphocytic choriomeningitis virus peptide gp61 conjugated to keyhole limpet hemocyanin (gp61-KLH) (Fig. 7e). Among SMARTA T cells, the frequency of TFH cells generated in the absence of Foxp1 was almost as high as that among T cells overexpressing Bcl-6 (Fig. 7e). Thus, in both the OT-II model system and the SMARTA model system, our results showed that Foxp1 exhibited profound dominance in regulating TFH cell differentiation.
Discussion
During the initial days of an immune response, how TFH cells arise from activated CD4+ T cells is still poorly understood3,4. Our study has established that Foxp1, through the use of two isoforms as a 'double-check' mechanism, is a rate-limiting and critical negative regulator of TFH cell differentiation. Our results have demonstrated important roles for the constitutive Foxp1A isoform and TCR-induced Foxp1D isoform in the control of the kinetics and magnitude of TFH cell development, which in turn greatly affects the subsequent GC and antibody responses.
Foxp1 has an essential role in maintaining the quiescence of naive T cells, partly by negatively regulating the pathway of the kinases MEK and Erk36. Signaling via MEK-Erk has a critical role in inducing ICOS expression by TCR stimulation: constitutively active MEK2 amplifies transcription of Icos, and a site in the promoter of Icos that is sensitive to Erk signaling has also been identified42. The initially enhanced induction of ICOS expression in Foxp1-deficient CD4+ T cells both in vitro and in vivo at early stages of T cell activation was probably due to the lack of the negative regulation of Foxp1 on MEK-Erk signaling. The enhanced expression of ICOS on Foxp1-deficient T cells, presumably through the interaction of ICOS with ICOSL-expressing DCs during the initial priming stage14, would lead to enhanced ICOS signaling, reflected by increased activation and degradation of Foxo1 protein, as we observed in ex vivo OT-II Foxp1-cKO T cells. It is plausible that in a T cell–dependent immune response, in the absence of Foxp1, higher ICOS expression and stronger ICOS signaling would allow more CD4+ T cells to differentiate into TFH cells. Meanwhile, the partial resistance of Foxp1-deficient CD4+ T cells to the blockade of ICOSL also suggested that mechanisms other than ICOS signaling contributed to the 'preferential' TFH differentiation of Foxp1-deficient CD4+ T cells and would provide a rationale for how Foxp1-deficient CD4+ T cells still differentiated into TFH cells in the absence of B cells. The Foxp1-mediated initial restraint of the expression of ICOS, a costimulator for T cell activation, could also be viewed as part of the Foxp1 function in controlling T cell quiescence. Thus, the function of the Foxp1 pathway in T cell quiescence seems to be linked to its role in TFH cell differentiation.
Although antigen dose has been shown to be important for TFH cell responses17,43, studies have suggested that the effector pattern of CD4+ T cells is also influenced by both the density and the dwell time of complexes of peptide and major histocompatibility complex class II rather than by TCR affinity alone44. OT-II and SMARTA model systems have been shown to generate different effector-cell patterns44. However, in both models, we found that deletion of Foxp1 resulted in an increased frequency of TFH cells, which demonstrates a critical role for Foxp1 in TFH cell differentiation. Also, in the OT-II and SMARTA model systems, deletion of Foxp1 in CD4+ T cells resulted in TFH cell frequency equal or higher than that achieved by overexpression of Bcl-6. The expression of mRNA encoding Bcl-6 in Foxp1-deficient TFH cells ex vivo at day 5 after immunization may have seemed to be slightly higher than that in control wild-type TFH cells. However, this could have been mainly due to the higher frequency of GC TFH cells (which are Bcl-6hi) in the Foxp1-deficient CXCR5+PD-1+ cell population at the time point of the experiments. At the protein level, we found that Foxp1-deficient and wild-type TFH cells expressed similar amounts of Bcl-6. Thus, we conclude that while Bcl-6 exerts essential positive control, Foxp1 is a crucial negative regulator of TFH cell differentiation.
TFH cell development may not require IL-21; however, in various model systems, IL-21 has been shown to have cell-intrinsic effects on TFH cell differentiation7,8,25. Extensive studies have shown that IL-21 acts directly on B cells and affects many aspects of GC B cell responses, including proliferation, survival and affinity selection, as well as differentiation into memory and plasma cells24,25,26. In our study, we found that IL-21 was a direct target of Foxp1 and that Foxp1 negatively regulated IL-21 in CD4+ T cells. Whereas it is very likely that the increased IL-21 production in Foxp1-deficient CD4+ T cells helped to generate the enhanced TFH cell responses and subsequent GC B cell responses we observed in our study, to what extent and whether Foxp1 may regulate some other TFH cell functions in helping B cell responses remains to be explored.
In our study, we found that the regulation of TFH cell differentiation by Foxp1 was dose dependent. The sum of Foxp1A and Foxp1D together served as a rate-limiting factor for the generation of TFH cells. In TFH cell differentiation, Foxp1A and 1D functioned in a very similar manner in that they both dampened such differentiation. The only domain that Foxp1D lacks is a polyglutamine-repeat domain, whose function has been characterized as the mediation of protein-protein interactions45. At present, little is known about partners that interact with Foxp1A or Foxp1D in their transcriptional complexes. Nevertheless, it is notable that Foxp1D, which peaked around day 2 in the OT-II OVA model, efficiently blocked TFH cell differentiation during the initial stage of the immune response. Such a 'gatekeeper' function for Foxp1D would be intrinsic to the immune response, as Foxp1D is induced mainly by TCR stimulation. Furthermore, our results showed that the 'preferential' TFH differentiation of Foxp1-deficient CD4+ T cells occurred at the expense of non-TFH cells. Thus, whereas this regulatory step is clearly critical for T cell–dependent GC responses, these results also indicate that Foxp1 may have roles in aspects of an immune response that depend on CD4+ non-TFH helper T cells as well.
In summary, our study has defined an important role for Foxp1 in TFH cell differentiation. Through the use of two isoforms as a 'double-check' mechanism, Foxp1 is a rate-limiting and critical negative regulator of TFH cell differentiation. The unique attributes of Foxp1 in CD4+ T cells may provide a useful pathway for manipulating humoral responses in vaccine development or the treatment of autoimmune disorders.
Methods
Mice.
All animals were maintained in specific pathogen–free barrier facilities and were used in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Wistar Institute. C57BL/6 mice were from the National Cancer Institute. Cre-ERT2+, RosaYFP, OT-II and μMT mice were from Jackson Laboratories. Mice with transgenic expression of Cd4-Cre and Ly5.1+ (CD45.1) C57BL/6 congenic mice were from Taconic. Foxp1f/f mice were backcrossed with C57BL/6 mice for 12 generations. Foxp1f/f mice were bred with Cre-ERT2+, RosaYFP and OT-II mice to generate OT-II Foxp1f/fRosaYFP, OT-II Foxp1f/+Cre-ERT2+RosaYFP and OT-II Foxp1f/fCre-ERT2+RosaYFP mice. Foxp1f/f mice were also bred with SMARTA mice to generate SMARTA Foxp1f/f mice. Mice with conditional transgenic expression of Foxp1A (Foxp1aTg/+) or Foxp1D (Foxp1dTg/+) were generated though the use of a Rosa26 knock-in approach46 and were backcrossed with C57BL/6 mice for at least seven generations. Foxp1aTg/+ and Foxp1dTg/+ mice were crossed with Cd4-Cre mice to generate Foxp1aTg/TgCd4-Cre and Foxp1dTg/TgCd4-Cre mice, respectively. Foxp1dTg/TgCd4-Cre mice were crossed with OT-II mice to generate OT-II Foxp1dTg/TgCd4-Cre mice. Foxp1aTg/TgCd4-Cre mice were crossed with Foxp1f/f and OT-II mice to generate OT-II Foxp1aTg/+Foxp1f/f Cd4-Cre mice. B1-8hi mice (which have transgenic expression of a B cell antigen receptor)40 were from G.D. Victora. SMARTA mice (which have transgenic expression of a TCR specific for specific for the lymphocytic choriomeningitis virus epitope of glycoprotein amino acids 66–77) were from S. Crotty and E.J. Wherry.
Flow cytometry, cell sorting and intracellular staining.
These procedures were done as described36. The sorted populations were >98% pure. Antibodies were as follows: phycoerythrin–anti-human CD271 (hNGFR; C40-1457), phycoerythrin–anti-CD44 (IM7), phycoerythrin–anti-CD69 (H1.2F3), phycoerythrin–anti-IL17A (TC11-18H10.1), phycoerythrin–anti-CD84 (mCD84.7), phycoerythrin–anti-CD45.2 (104), peridinin chlorophyll protein–cyanine 5.5–anti-Vα2 (B20.1), peridinin chlorophyll protein–cyanine 5.5–anti-CD25 (PC61), peridinin chlorophyll protein–cyanine 5.5–anti-CD45.2 (104), allophycocyanin–anti-CD62L (MEL-14), allophycocyanin–anti-CD45.1 (A20), allophycocyanin–anti-ICOS (C398.4A), allophycocyanin–anti-CD4 (GK1.5), phycoerythrin–indotricarbocyanine–anti-CD4 (GK1.5), phycoerythrin–indotricarbocyanine–anti-PD1 (29F.1A12) and Brilliant Violet 421–anti-IL4 (11B11; all from BioLegend); phycoerythrin–anti-Ly108 (eBio13G3-19D), peridinin chlorophyll protein–cyanine 5.5–anti-interferon-γ (XMG1.2), peridinin chlorophyll protein–cyanine 5.5–anti-B220 (RA3-6B2), allophycocyanin–anti-BTLA (8F4 ), allophycocyanin–eFluor 780–anti-CD4 (RM4-5) and Alexa Fluor 647–anti-CD200 (OX9; all from eBioscience); phycoerythrin–anti-GL7 (GL7) and purified or biotinylated rat anti–mouse CXCR5 (2G8; both from BD Biosciences); biotinylated goat anti–rat IgG (112-065-062; Jackson ImmunoResearch Laboratories); and Brilliant Violet 421–streptavidin (BioLegend). Rabbit polyclonal anti-Foxp1 (generated by AbMART) recognizes a carboxy-terminal epitope shared by all four Foxp1 isoforms34,36. Fluorescein isothiocyanate–labeled lectin was from Sigma. Dead cells were excluded through the use of a Live/Dead Fixable Aqua Dead Cell staining kit (Invitrogen). Intracellular IL-21 was detected with a chimera of recombinant mouse IL-21 receptor and the Fc fragment (R&D Systems), followed by Alexa Fluor 647–conjugated anti–human IgG (109-605-098; Jackson ImmunoResearch Laboratories).
For intracellular staining of Foxp1, Foxo1 or Bcl-6, cells were fixed with 4% formaldehyde after staining of cell surface markers (antibodies identified above) and were permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS). Cells were stained overnight at 4 °C with rabbit anti-Foxp1 (identified above), rabbit anti-Foxo1 (C29H4; Cell Signaling Technology) or phycoerythrin–anti-Bcl-6 (K112-91; BD Biosciences). For reduction of background, cells to be stained for Foxp1 were washed three times for 5 min each (with rotation) with 0.01% Triton X-100 in PBS after incubation with anti-Foxp1, then were stained with Alexa Fluor 647–labeled goat antibody to rabbit immunoglobulin (A-21244; Invitrogen).
For intracellular staining of cytokines, cells were stimulated for 4 h with PMA (phorbol 12-myristate 13-acetate; 5 ng/ml) and ionomycin (0.5 μg/ml), and the staining procedures were done as described36.
T cell stimulation and retroviral transduction.
For in vitro activation of T cells, purified naive CD4+ T cells were stimulated for 48 h with anti-CD3 (0.5 μg/ml; 145-2C11; eBioscience) and anti-CD28 (1 μg/ml; 37.51; eBioscience) in plates precoated with goat antibody to hamster IgG (0.3 mg/ml; 55397; MP Biomedicals) in complete T cell medium (Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, penicillin-streptomycin, nonessential amino acids, sodium pyruvate, vitamins, 10 mM HEPES and 50 μM 2-mercaptoethanol), then their populations were expanded for another 2–3 d in T cell medium containing 100 U/ml recombinant human IL-2.
The plasmids MigR1-Bcl-6-GFP and MigR1-Cre-GFP were gifts from M.E. Pipkin and N.A. Speck, respectively. The open reading frames of Bcl6 were also subcloned into the retroviral vector MSCV-IRES-hNGFR (a gift from W. Pear). Retrovirus containing sequence encoding Bcl-6, Foxp1A or Cre was produced in human embryonic kidney HEK293T cells (American Type Culture Collection) by cotransfection with retroviral vectors and helper plasmids.
For transduction of retrovirus, purified naive CD4+ T cells were stimulated for about 28 h with anti-CD3 and anti-CD28 as described above. Cells were transduced with virus-containing medium supplemented with polybrene (6 μg/ml) and were centrifuged for 2 h at 650g. After 20 h of culture, the culture medium was replaced with complete T cell medium supplemented with 100 U/ml recombinant human IL-2, followed by incubation for 2 d.
CellTrace labeling.
Purified CD44loVα2hi naive CD4+ T cells were washed twice with PBS and were incubated for 20 min at 37 °C at a density of 1 × 107 cells per ml in PBS with 2 μM CellTrace (CellTrace Violet Cell Proliferation Kit; Invitrogen), then washed with DMEM medium with 10% FBS. Labeled CD4+ T cells were stimulated with anti-CD3 and anti-CD28 (both identified above) or were transferred into Ly5.1+ C57BL/6 mice (0.25 × 106 cells per mouse).
Adoptive transfer.
Six- to ten-week old mice were treated daily for 4 d with tamoxifen (Sigma-Aldrich) at a dose of 1.5 mg per mouse and were allowed to 'rest' for 1 d. Samples were enriched for CD4+ T cells by negative selection with magnetic beads (CD4+ T cell Isolation Kit II; Miltenyi Biotec). YFP+ CD44loVα2hi CD4+ naive T cells from OT-II Foxp1f/fCre-ERT2+RosaYFP, OT-II Foxp1f/+Cre-ERT2+RosaYFP and OT-II Foxp1+/+Cre-ERT2+RosaYFP mice, or YFP− CD44loVα2hi CD4+ naive T cells from OT-II Foxp1f/fRosaYFP were further sorted with a MoFlow cell sorter (DakoCytomation). In experiments in which OT-II Foxp1aTg/+Foxp1f/fCd4-Cre T cells were transferred, 4- to 5-week-old mice were used. After being washed with PBS, 0.25 × 106 sorted naive cells were transferred into the recipient mice by injection into the tail vein, followed by immunization by intraperitoneal injection of 50 μg NP14-OVA (Biosearch Technologies) precipitated in alum adjuvant (Pierce). For the transfer of activated T cells, 1 × 106 purified cells were transferred into recipient mice. Then, 1–3 d later, mice were immunized by intraperitoneal injection of 100 μg NP14-OVA in alum (for OT-II cells) or 10 μg gp61-KLH (for SMARTA cells).
For in vivo blockade of ICOSL, recipient mice were treated with 500 μg monoclonal antibody to ICOSL (HK5.3; Bio X Cell) or PBS by intraperitoneal injection every other day starting at day 0 before immunization. The extent of blockade was calculated as (N1−N2) / N1, where 'N1' is the frequency of TFH cells without antibody blockade (PBS) and 'N2' is the frequency of TFH cells after treatment with anti-ICOSL.
Infection with influenza virus.
Mice were immobilized with ketamine and xylazine (70 mg and 10 mg, respectively, per kg body weight) and were infected intranasally with mouse-adapted influenza virus strain A/Puerto Rico/8/34 (PR8) (H1N1, Mount Sinai strain) at a dose of 450 TCID50 (half-maximal tissue culture infectious dose) per 30 μl.
TH1, TH2 and TH17 differentiation in vitro.
YFP+ and YFP− naive T cells were sorted from tamoxifen-treated Foxp1f/fCre-ERT2+RosaYFP and Foxp1f/fRosaYFP mice, respectively, and were stimulated for 2 d with anti-CD3 and anti-CD28 (both identified above) in medium supplemented as follows: for TH1 differentiation, 10 ng/ml IL-12 (R&D Systems) and 10 μg/ml anti-IL-4 (11B11; eBioscience); for TH2 differentiation, 10 ng/ml IL-4 (R&D Systems), 10 μg/ml anti-IL-12 (C17.8; eBioscience) and 10 μg/ml anti-interferon-γ (XMG1.2; BioXcell); for TH17 differentiation, 1 ng/ml TGF-β (R&D Systems), 20 ng/ml IL-6 (R&D Systems), 10 μg/ml anti-interferon-γ (identified above) and 10 μg/ml anti-IL-4 (identified above). Cells were then cultured in T cell medium supplemented with 100 U/ml recombinant human IL-2 under TH1-, TH2- or TH17-polarizing conditions. Cells were stimulated for 4 h with PMA (5 ng/ml) and ionomycin (0.5 μg/ml) and then intracellular cytokines were stained (antibodies identified above).
Immunoblot analysis.
Cells were washed with PBS twice and then were lysed in RIPA buffer containing 1× Protease Inhibitor Cocktail (Roche). Cell lysates were separated by SDS-PAGE and then anayzed by immunoblot with rabbit anti-Foxp1 (identified above), mouse anti-Bcl-6 (K112-91; BD Biosciences) or goat anti-β-actin (I-19; Santa Cruz Biotechnology).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation of Foxp1 in wild-type or Foxp1-cKO CD4+ T cells activated under TH1 conditions was done as described36. Precipitated DNA and input DNA were assessed by quantitative real-time PCR with SYBR Green PCR Master Mix (Applied Biosystems). The sequences of the primer pairs used were as follows: for the Il21 promoter region, forward, 5′-AGGGATGGATAGAGTCCACAA-3′, and reverse, 5′-GCTGCTTTACTCATTGCAGAAG-3′; and for the Il21 control region, forward, 5′-GCAGTAAGGGAAGAAGGTCAAG-3′, and reverse, 5′-GGGCTGGATTTGTGGAAAGA-3′.
Real-time RT-PCR.
Purified naive CD4+ T cells from 6- to 8-week-old Foxp1f/fRosaYFP and Foxp1f/fCre-ERT2+RosaYFP mice were stimulated with anti-CD3 and anti-CD28 (both identified above) in T cell medium with recombinant human IL-2 (10 U/ml) under TFH cell–like conditions as follows: 20 ng/ml IL-6, 20 ng/ml IL-21 (R&D Systems), 10 μg/ml anti-IL-4 (identified above) and 10 μg/ml anti-interferon-γ (identified above). Some T cells were cultured under TH1-polarizing conditions with recombinant human IL-2 (10 U/ml) for 4 d. Total RNA was purified as described36. Expression of mRNA was normalized to Rpl32 expression and presented as relative to wild-type CD4+ T cells. The primers for analysis of TFH cell–related gene expression by real-time RT-PCR were as follows: for Bcl6 (forward, 5′-GTGATGACCACAGCCATGTACCTGC-3′, and reverse, 5′-CACGACCTCGGTAGGCCATGATG-3′), for cMaf (forward, 5′-AGCAGTTGGTGACCATGTCG-3′, and reverse, 5′-TGGAGATCTCCTGCTTGAGG-3′), for Prdm1 (forward, 5′-CTTG TGTGGTATTGTCGGGAC-3′, and reverse, 5′-CACGCTGTACTCTCTCTTGG-3′), for Tbx21 (forward, 5′-CAACAACCCCTTTGCCAAAG-3′, and reverse, 5′-TCCCCCAAGCAGTTGACAGT-3′), for Irf4 (forward, 5′-CTTTGAGGAATTGGTCGAGAGG-3′, and reverse, 5′-GAGAGCCATAAGGTGCTGTCA-3′), for Baf (forward, 5′-AGCCGACAGAGACAGACACAGAAA-3′, and reverse, 5′-TCCTCGGTGAGCTCTTTGATCTCT-3′), for Sh2d1a (forward, 5′-GAAACAGGTTCTTGGAGTGCC-3′, and reverse, 5′-GTCACGATGCCTTGATCCG-3′), for Icos (forward, 5′-CTCACCAAGACCAAGGGAAGC-3′′, and reverse, 5′-CCACAACGAAAGCTGCACACC-3′), for Il6r (forward, 5′-GGT GGCCCAGTACCAATGC-3′, and reverse, 5′-GGACCTGGACCACGTGCT-3′), for Il21r (forward, 5′-TTTCACGGCCTCCAGCATAGAGTT-3′, and reverse, 5′-ACCAGGCTCAGACATTCCATCACA-3′), for Cd40lg (forward, 5′-GTGAGGAGATGAGAAGGCAA-3′, and reverse, 5′-CACTGTAGAACGGATGCTGC-3′), for Foxp1 (forward, 5′-CTGAATCTGGTATCAAGTGTCACCCTCT-3′, and reverse, 5′-GATTCGAGAATGGCCTGCCTGA-3′), and for Rpl32 (forward, 5′-CCCAACATCGGTTATGGGAGCA-3′, and reverse, 5′-GATGGCCAGCTGTGCTGC-3′).
Microarray.
For sample preparation and hybridization, total RNA from purified CXCR5hiPD1hi TFH or CXCR5−PD1− non-TFH cells was isolated with TRIzol reagent according to the manufacturer's recommendations (Invitrogen). RNA quality was assesse with a Bioanalyzer (Agilent). Only samples with RNA-integrity numbers of ≥9.5 were used for further studies. Equal amounts (400 ng) of total RNA was amplified as recommended by Illumina and was hybridized to the Illumina MouseWG 6v2 mouse whole-genome bead arrays.
For data preprocessing, Illumina GenomeStudio software was used to export expression values and the calculated detection P values for each probe of each sample. Signal-intensity data were log2 transformed and quantile-normalized. Only genes with a significant detection P value (P < 0.05) in at least one of six samples were considered. The data were submitted to the GEO database.
For the external data set, preprocessed external sets of TFH cell–related data with accession numbers GSE24574, GSE16697 and GSE40068 were downloaded from the GEO database9,16,47.
For analysis of differences in expression, the SAM ('significance analysis of microarrays') method48 was used to find genes expressed differently in the TFH cell class (two replicates) and non-TFH cell class (one replicate) with the 'One class' option for data from TFH cells, with expression values for non-TFH sample subtracted from those values. Genes expressed differently in the data sets GSE24574, GSE16697 and GSE40068 were identified by the two-tailed Student's t-test, and the false-discovery rate was estimated with a published procedure49. Genes with a false-discovery rate of <5% were considered significant.
To identify overlapping of genes encoding TFH cell markers in our Illumina data and in the publicly available Affymetrix data in GSE24574, GSE16697 and GSE40068, we used accession codes of the Entrez databases (National Center for Biotechnology Information) associated with each Illumina or Affymetrix probe as identified by software of the DAVID bioinformatics database (Database for Annotation, Visualization and Integrated Discovery)50. For Entrez accession codes with multiple associated probes, only the probe with the highest expression was considered.
For the identification of TFH cell markers, a set of genes with significant expression (false-discovery rate, <5%) in GSE24574, GSE16697 and GSE40068 and nominal P value of <0.01 in wild-type samples were 'called' as TFH cell markers; these were used to estimate similarity between OT-II Foxp1-WT cells and OT-II Foxp1-cKO TFH cells.
Enzyme-linked immunospot assay.
ELISPOT plates (Millipore) were coated overnight at 4 °C with 10 μg/ml NP25-BSA (Biosearch Technologies) or BSA in PBS, then nonspecific binding was blocked with DMEM medium containing 10% FBS. Serial dilutions of splenocytes were cultured in the coated plates at 37 °C in 5% CO2. After incubation overnight, plates were washed with PBS containing 0.05% Tween 20 and were incubated with alkaline phosphatase–conjugated anti-IgM (1020-04; SouthernBiotech), anti-IgG (1030-04; SouthernBiotech) and anti-IgG1 (115-055-205; Jackson ImmunoResearch Laboratiories). Antibody spots were developed with the NBT/BCIP substrate (Santa Cruz). The frequency of antibody-secreting cells was determined with an ImmunoSpot Reader (CTL) and ImmunoSpot satellite software (CTL).
Enzyme-linked immunosorbent assay.
ELISA plates (Costa) were coated overnight at 4 °C with 10 μg/ml NP4-BSA (Biosearch Technologies, Inc.) or NP25-BSA in 0.1 M carbonate buffer (pH 9.0). Nonspecific binding in the coated plates was blocked with 3% BSA in PBS and plates containing serial dilutions of serum were incubated for 2 h at room temperature, followed by incubation with alkaline phosphatase–coupled anti-IgG (identified above). Alkaline phosphatase activity was visualized with p-nitrophenyl phosphate substrate (Sigma-Aldrich) and the absorbance at 405 nm was determined with an ELISA reader (Molecular Devices). Titers represent the highest serum dilution with a value of ≥0.1 above background for absorbance at 405 nm.
Histology.
These procedures were done as described40. 0.1 × 106 purified naive OT-II Foxp1-WT or OT-II Foxp1-cKO T cells were transferred together with 0.5 × 106 B1-8hi B cells into Ly5.1+ SMARTA mice, followed by immunization of the recipient mice with NP-OVA in alum. At day 6 after immunization, spleens were fixed for 1 h at 4 °C in 4% paraformaldehyde and 10% sucrose in PBS, then were incubated overnight in 30% sucrose before being embedded in optimum cutting temperature compound and cryosectioned. Samples were then fixed for 10 min at −20 °C in acetone, nonspecific binding was blocked with a Streptavidin/Biotin Blocking Kit (Vector Labs) and samples were stained in the following three steps: first, with purified rat anti-mouse (mAID-2; eBioscience) plus biotin–anti-CD45.2 (104; BD Biosciences); second, with Alexa Fluor 555–conjugated goat polyclonal anti-rat (A-21434; Invitrogen) plus Alexa Fluor 488–streptavidin (Invitrogen); and third, with Alexa Fluor 647–conjugated rat antibody to mouse IgD (11-26; BioLegend). All samples were incubated in a solution of 5% BSA, 10% normal mouse serum and 0.1% Triton X-114 in PBS. Mounted sections were imaged on a Zeiss 700 confocal microscope with a 5× objective with a numerical aperture of 0.25 and a 20× objective with a numerical aperture of 0.8.
Statistics.
Two-tailed Student's t-tests and paired t-tests (Supplementary Table 2) were used for calculation of P values, except for calculation of P values for microarray analyses, in which SAM test was also used.
Accession codes.
GEO: microarray data, GSE50725.
References
Breitfeld, D. et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192, 1545–1552 (2000).
Schaerli, P. et al. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 192, 1553–1562 (2000).
Vinuesa, C.G., Tangye, S.G., Moser, B. & Mackay, C.R. Follicular B helper T cells in antibody responses and autoimmunity. Nat. Rev. Immunol. 5, 853–865 (2005).
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011).
Ansel, K.M., McHeyzer-Williams, L.J., Ngo, V.N., McHeyzer-Williams, M.G. & Cyster, J.G. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J. Exp. Med. 190, 1123–1134 (1999).
Chtanova, T. et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173, 68–78 (2004).
Vogelzang, A. et al. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity 29, 127–137 (2008).
Nurieva, R.I. et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29, 138–149 (2008).
Johnston, R.J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009).
Nurieva, R.I. et al. Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 (2009).
Yu, D. et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31, 457–468 (2009).
Cannons, J.L. et al. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity 32, 253–265 (2010).
Kerfoot, S.M. et al. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity 34, 947–960 (2011).
Choi, Y.S. et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34, 932–946 (2011).
Goenka, R. et al. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J. Immunol. 187, 1091–1095 (2011).
Liu, X. et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 209, 1841–1852 (2012).
Deenick, E.K. et al. Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity 33, 241–253 (2010).
Xu, H. et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature 496, 523–527 (2013).
Choi, Y.S., Eto, D., Yang, J.A., Lao, C. & Crotty, S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J. Immunol. 190, 3049–3053 (2013).
Johnston, R.J., Choi, Y.S., Diamond, J.A., Yang, J.A. & Crotty, S. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 209, 243–250 (2012).
Ballesteros-Tato, A. et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36, 847–856 (2012).
Schmitt, N. et al. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity 31, 158–169 (2009).
Ma, C.S. et al. Early commitment of naive human CD4+ T cells to the T follicular helper (TFH) cell lineage is induced by IL-12. Immunol. Cell Biol. 87, 590–600 (2009).
Ozaki, K. et al. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 173, 5361–5371 (2004).
Linterman, M.A. et al. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 207, 353–363 (2010).
Zotos, D. et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 207, 365–378 (2010).
Bauquet, A.T. et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10, 167–175 (2009).
Kwon, H. et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31, 941–952 (2009).
Ise, W. et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12, 536–543 (2011).
Bollig, N. et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc. Natl. Acad. Sci. USA 109, 8664–8669 (2012).
Liu, X. et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 507, 513–518 (2014).
Carlsson, P. & Mahlapuu, M. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250, 1–23 (2002).
Calnan, D.R. & Brunet, A. The FoxO code. Oncogene 27, 2276–2288 (2008).
Wang, B., Lin, D., Li, C. & Tucker, P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J. Biol. Chem. 278, 24259–24268 (2003).
Feng, X. et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood 115, 510–518 (2010).
Feng, X. et al. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 12, 544–550 (2011).
Yusuf, I. et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol. 185, 190–202 (2010).
Linterman, M.A. et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17, 975–982 (2011).
Chung, Y. et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 17, 983–988 (2011).
Shulman, Z. et al. T follicular helper cell dynamics in germinal centers. Science 341, 673–677 (2013).
Gigoux, M. et al. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA 106, 20371–20376 (2009).
Tan, A.H., Wong, S.C. & Lam, K.P. Regulation of mouse inducible costimulator (ICOS) expression by Fyn-NFATc2 and ERK signaling in T cells. J. Biol. Chem. 281, 28666–28678 (2006).
Baumjohann, D. et al. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity 38, 596–605 (2013).
Tubo, N.J. et al. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153, 785–796 (2013).
Schaefer, M.H., Wanker, E.E. & Andrade-Navarro, M.A. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 40, 4273–4287 (2012).
Xiao, C. et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159 (2007).
Kitano, M. et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 34, 961–972 (2011).
Zhang, S. A comprehensive evaluation of SAM, the SAM R-package and a simple modification to improve its performance. BMC Bioinformatics 8, 230 (2007).
Storey, J.D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank K. Rajewsky (Max Delbruck Center for Molecular Medicine) for the Rosa26 targeting vector; E.J. Wherry (University of Pennsylvania) for the SMARTA mice; M.E. Pipkin (The Scripps Research Institute) for the plasmid MigR1-Bcl-6-GFP; N.A. Speck (University of Pennsylvania) for the plasmid MigR1-Cre-GFP; W. Pear (University of Pennsylvania) for the plasmid MSCV-IRES-hNGFR; K. Mozdzanowska and J. Hayden for technical help with histology; J.S. Faust, D.E. Ambrose and S. Weiss for technical help with flow cytometry; M.S. Wright, M. Houston-Leslie and D. DiFrancesco for help at the Animal Facility of the Wistar Institute; and P. Wickramasinghe for help with microarray data and accession codes. Supported by the US National Institutes of Health (AI095439 and AI103162 to H.H.; CA132098 and 1S10RR024693 to L.C.S.; CA31534 to H.O.T.; AI083022 to J.E.; 5DP5OD012146 to G.D.V.; and AI063107 to S.C.), the Alliance for Cell Gene Therapy Foundation (H.H.), the Wawa Foundation (H.H.), the Martha W. Rogers Trust (H.H.), and the Wistar Cancer Center (P30 CA10815).
Author information
Authors and Affiliations
Contributions
H.W. designed and did experiments; J.G., X.W., E.B., A.I.W., J.T., Y.S.C., H.T., T.J.D., L-Y.C., S.L.S., E.K.B., J.W., L.T. and X.W. helped with experiments; C.X. and P.J. helped generate mice with conditional transgenic expression of Foxp1A and Foxp1D; S.C. provided gp61-KLH and supervised ICOSL-blocking experiments; G.D.V. supervised the immunohistochemistry staining and confocal microscopy; H.O.T. supervised generation of Foxp1f/f mice and provided them; J.E. supervised the viral infection experiment; A.V.K. did bioinformatics analysis under the supervision of L.C.S.; H.H. designed experiments and provided overall direction; and H.W., J.G., A.V.K., A.I.W., T.J.D., H.O.T., J.E. and H.H. wrote the paper. Authors who contributed equally to this work are listed in alphabetical order.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Foxp1D induction by TCR stimulation in vitro and in vivo.
(a) Wild-type naive CD4+ T cells were stimulated by plate-bound anti-CD3 antibodies at different concentrations for 48 hours, then analyzed by immunoblotting for Foxp1 expression and by staining for cell surface markers CD44. (b) Wild-type naïve OT-II T cells were transferred into Ly5.1+ C57BL/6 recipient mice followed by immunization with NP-OVA in alum. Foxp1 expression was analyzed by intracellular staining in donor OT-II T cells at different time points after immunization. Foxp1f/fCd4-Cre (KO) CD4+ T cells were used as controls. Results represent three (a) and two (b) independent experiments.
Supplementary Figure 2 TFH cell differentiation of Foxp1-deficient OT-II T cells in Ly5.1+ C57BL/6 recipient mice.
Purified naive OT-II Foxp1-WT or OT-II Foxp1-cKO T cells were transferred into Ly5.1+ C57BL/6 recipient mice followed by immunization with NP-OVA in alum. (a) Percentages of PD-1hiCXCR5hi TFH cells and GL7hiCXCR5hi GC TFH cells in splenic donor OT-II T cells 6 days after immunization. (b) Bcl-6 expression in donor OT-II TFH cells by intracellular staining 6 days after immunization. (c) Phenotypes of donor OT-II TFH cells by cell surface staining 6 days after immunization. Results represent at least two independent experiments.
Supplementary Figure 3 The effect of Foxp1 on TFH differentiation is cell intrinsic.
(a) Purified naïve OT-II Foxp1-WT or OT-II Foxp1-cKO T cells were transferred into Ly5.1+ SMARTA recipient mice followed by immunization with NP-OVA in alum. Foxp3 expression in splenic donor OT-II T cells and endogenous CD4+ T cells in SMARTA recipient mice were analyzed 6 days after immunization. (b) Naïve OT-II Foxp1-WT and OT-II Foxp1-cKO T cells were mixed at 1:1 ratio and co-transferred into Ly5.1+C57BL/6 recipient mice followed by immunization with NP-OVA in alum. The percentage of TFH cells in donor OT-II T cells in the mLN of the recipient mice were analyzed 5 days after immunization. Results represent two independent experiments.
Supplementary Figure 4 TH1, TH2 and TH17 differentiation of Foxp1-deficient CD4+ T cells.
Purified naïve CD4+T cells from tamoxifen-treated Foxp1f/fRosaYFPmice (WT) or Foxp1f/fCre-ERT2+RosaYFPmice (cKO) were stimulated by anti-CD3 and anti-CD28 antibodies under TH1-, TH 2- or TH 17-polarizing culture conditions for 2 days, followed by expansion in T cell medium with IL-2 for two more days. Cytokines were examined by intracellular staining. Results represent three independent experiments.
Supplementary Figure 5 Generation of mice with conditional transgenic expression of Foxp1A and Foxp1D.
(a) Schematic diagram of the targeting strategy for Foxp1A (Foxp1aTg/+) or Foxp1D (Foxp1dTg/+) conditional transgenic mice. (b) Southern blotting analyses of Foxp1aTg/+ and Foxp1dTg/+ mice. (c) Expression of cell surface markers and GFP in Foxp1aTg/TgCd4-Cre CD4+ T cells. (d) Expression of cell surface markers and GFP in Foxp1dTg/TgCd4-Cre CD4+ T cells. Results represent at least two independent experiments.
Supplementary Figure 6 Both Foxp1A and Foxp1D negatively regulate TFH cell development.
(a, b) Foxp1aTg/TgCd4-Cre mice were infected with influenza virus PR8. The percentages and numbers of PD-1hiCXCR5hi TFH cells (a) and GC B cells (b) in the spleens of the infected mice were analyzed. Foxp1aTg/Tg mice were used as controls. Error bars represent s.d. (n=5). (c, d) Foxp1dTg/TgCd4-Cre mice were infected with PR8 virus. The percentages and numbers of PD-1hiCXCR5hi TFH cells (c) and GC B cells (d) in the spleens of the infected mice were analyzed. Foxp1dTg/Tg mice were used as controls. Error bars represent s.d. (n=4). Results represent at least two independent experiments. *P < 0.05, **P < 0.01.
Supplementary Figure 7 Transgenic expression of Foxp1D suppresses TFH cell differentiation but not the general CD4+ T cell responses.
(a) Purified naive CD4+ T cells from OT-IIFoxp1dTg/TgCd4-Cre or control OT-IIFoxp1dTg/Tg mice were transferred into Ly5.1+ C57BL/6 recipient mice followed by immunization with NP-OVA in alum. Seven days after immunization, the percentages of PD-1hiCXCR5hi TFH cells in splenic donor OT-II T cells were analyzed. (b) Purified naive CD4+ T cells from OT-IIFoxp1dTg/TgCd4-Cre or control OT-IIFoxp1dTg/Tg mice were mixed at 1:1 ratio and co-transferred into Ly5.1+ C57BL/6 recipient mice followed by immunization with NP-OVA in alum. The ratios of GFP+ Foxp1D transgenic OT-II T cells to control OT-II and the percentages of TFH cells in splenic donor OT-II T cells were analyzed 7 days after immunization. Results represent two independent experiments.
Supplementary Figure 8 Expression of TFH cell–related genes in wild-type and Foxp1-deficient TFH cell–like cells in vitro and Foxp1D specifically inhibits IL-21 production in vitro.
(a) Relative mRNA expression in wild-type and Foxp1-deficient T cells activated under TFH-like cell culture conditions in vitro. Error bars represent standard error of the mean (s.e.m.). (b) Wild-type (Foxp1dTg/Tg) or Foxp1D transgenic (Foxp1dTg/TgCd4-Cre) CD4+ T cells were stimulated by anti-CD3 and anti-CD28 antibodies in vitro under TH1-polarizing culture conditions for three days and expanded for additional 2 days in medium.IL-21 production was examined on day 5 by intracellular staining.
Supplementary Figure 9 Recovery of donor OT-II T cells at the initial stage of the responses in B cell–deficient μMT recipient mice versus B cell–intact recipient mice.
The day 3 data of total donor OT-II T cells in the spleens of the recipient mice of Fig. 7b and Fig. 2a are shown. Error bars represent s.d. (μMT recipients: n=2; SMARTA recipients: n=4). Results represent at least two independent experiments.
Supplementary Figure 10 TFH cell differentiation of Foxp1-deficient CD4+ T cells in μMT recipient mice.
Purified naïve OT-II Foxp1-WT and OT-II Foxp1-cKO T cells were transferred into μMT recipient mice followed by immunization with NP-OVA in alum. (a) Phenotypes of donor OT-II T cells in the spleens of the recipient mice 5 days after immunization. (b) Percentages of GL7hiCXCR5hi GC TFH cells in splenic donor OT-II T cells 4 and 7 days after immunization. (c) Percentages of IL-21-producing cells in day 5 splenic donor OT-II T cells by intracellular staining. Results represent at least two independent experiments.
Supplementary Figure 11 Retroviral overexpression of Bcl-6.
(a) Immunoblotting of Bcl-6 in HEK293T cells transfected with plasmid MSCV-IRES-hNGFR (Mock) or MSCV-Bcl-6-hNGFR. (b) Immunoblotting (left panel) or intracellular staining (right panel) of Bcl-6 in CD4+ T cells infected with RV-Ctrl or RV-Bcl-6 retroviruses. (c) Wild-type or Foxp1-deficient naïve OT-II T cells transduced with RV-Ctrl, RV-Bcl-6, or RV-Foxp1A were transferred into Ly5.1+ C57BL/6 recipient mice. Three days later, the recipient mice were immunized with NP-OVA in alum. TFH cell differentiation of hNGFR+ cells in the mLN was analyzed 5 days after immunization. Results represent at least two independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 and Supplementary Tables 1 and 2 (PDF 2649 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Geng, J., Wen, X. et al. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol 15, 667–675 (2014). https://doi.org/10.1038/ni.2890
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2890
This article is cited by
-
Spatiotemporal resolution of germinal center Tfh cell differentiation and divergence from central memory CD4+ T cell fate
Nature Communications (2023)
-
The phosphatase PTEN links platelets with immune regulatory functions of mouse T follicular helper cells
Nature Communications (2022)
-
Continuous activation of polymorphonuclear myeloid-derived suppressor cells during pregnancy is critical for fetal development
Cellular & Molecular Immunology (2021)
-
Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits
Nature Immunology (2020)
-
ZIKV infection induces robust Th1-like Tfh cell and long-term protective antibody responses in immunocompetent mice
Nature Communications (2019)