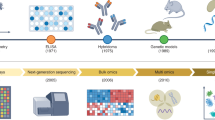
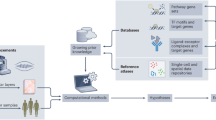
Abstract
The immune system is a highly complex and dynamic system. Historically, the most common scientific and clinical practice has been to evaluate its individual components. This kind of approach cannot always expose the interconnecting pathways that control immune-system responses and does not reveal how the immune system works across multiple biological systems and scales. High-throughput technologies can be used to measure thousands of parameters of the immune system at a genome-wide scale. These system-wide surveys yield massive amounts of quantitative data that provide a means to monitor and probe immune-system function. New integrative analyses can help synthesize and transform these data into valuable biological insight. Here we review some of the computational analysis tools for high-dimensional data and how they can be applied to immunology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Pascual, V., Chaussabel, D. & Banchereau, J. A genomic approach to human autoimmune diseases. Annu. Rev. Immunol. 28, 535–571 (2010).
Boisson, B. et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13, 1178–1186 (2012).
Chaussabel, D. et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 29, 150–164 (2008).
Querec, T.D. et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10, 116–125 (2009).
Nakaya, H.I. et al. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 12, 786–795 (2011).
Furman, D. et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol. Syst. Biol. 9, 659 (2013).
Obermoser, G. et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 38, 831–844 (2013).
Berry, M.P. et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 (2010).
Cliff, J.M. et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J. Infect. Dis. 207, 18–29 (2013).
Bloom, C.I. et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS ONE 7, e46191 (2012).
Law, G.L., Korth, M., Benecke, A. & Katze, M. Systems virology: host-directed approaches to viral pathogenesis and drug targeting. Nat. Rev. Microbiol. 11, 455–466 (2013).
Chiche, L., Jourde-Chiche, N., Pascual, V. & Chaussabel, D. Current perspectives on systems immunology approaches to rheumatic diseases. Arthritis Rheum. 65, 1407–1417 (2013).
Hummel, M. et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 354, 2419–2430 (2006).
Casanova, J.-L., Abel, L. & Quintana-Murci, L. Immunology taught by human genetics. Cold Spring Harb. Symp. Quant. Biol. 4, a007260 (2013).
Xavier, R.J. & Rioux, J.D. Genome-wide association studies: a new window into immune-mediated diseases. Nat. Rev. Immunol. 8, 631–643 (2008).
Orrù, V. et al. Genetic variants regulating immune cell levels in health and disease. Cell 155, 242–256 (2013).
Visscher, P.M., Brown, M., McCarthy, M. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Cho, J.H. & Gregersen, P. Genomics and the multifactorial nature of human autoimmune disease. N. Engl. J. Med. 365, 1612–1623 (2011).
Cotsapas, C. et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 7, e1002254 (2011).
Goris, A. & Liston, A. The immunogenetic architecture of autoimmune disease. Cold Spring Harb. Perspect. Biol. 4, a007260 (2012).
Voight, B.F. & Cotsapas, C. Human genetics offers an emerging picture of common pathways and mechanisms in autoimmunity. Curr. Opin. Immunol. 24, 552–557 (2012).
Bolze, A. et al. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science 340, 976–978 (2013).
Flomenberg, N. et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood 104, 1923–1930 (2004).
Spellman, S.R. et al. A perspective on the selection of unrelated donors and cord blood units for transplantation. Blood 120, 259–265 (2012).
Wang, C. et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc. Natl. Acad. Sci. USA 109, 8676–8681 (2012).
Trowsdale, J. The MHC, disease and selection. Immunol. Lett. 137, 1–8 (2011).
Prugnolle, F. et al. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 15, 1022–1027 (2005).
Newell, E.W et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat. Biotechnol. 31, 623–629 (2013).
DeKosky, B.J. et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 31, 166–169 (2013).
Jiang, N. et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 5, 171ra19 (2013).
Wu, X. et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011).
Wu, D. et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci. Transl. Med. 4, 134ra63 (2012).
Robins, H.S. et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med. 2, 47ra64 (2010).
Boyd, S.D. et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 1, 12ra23 (2009).
Jiang, N. et al. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc. Natl. Acad. Sci. USA 108, 5348–5353 (2011).
Bolotin, D.A. et al. MiTCR: software for T-cell receptor sequencing data analysis. Nat. Methods 10, 813–814 (2013).
Alamyar, E., Giudicelli, V., Li, S., Duroux, P. & Lefranc, M.-P. IMGT/HighV-QUEST: the IMGT® web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 882, 569–604 (2012).
Boyd, S.D. Diagnostic applications of high-throughput DNA sequencing. Annu. Rev. Pathol. 8, 381–410 (2013).
Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
Mortazavi, A., Williams, B., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Fairfax, B.P. et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet. 44, 502–510 (2012).
Zhang, J.A., Mortazavi, A., Williams, B., Wold, B. & Rothenberg, E. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149, 467–482 (2012).
Yosef, N. et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468 (2013).
Shalek, A.K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Robinson, M.D., McCarthy, D. & Smyth, G. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Li, J., Witten, D., Johnstone, I. & Tibshirani, R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics 13, 523–538 (2012).
Anders, S., Reyes, A. & Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Efron, B. & Tibshirani, R. On testing the significance of sets of genes. Ann. Appl. Stat. 1, 107–129 (2007).
Haining, W.N. & Wherry, E.J. Integrating genomic signatures for immunologic discovery. Immunity 32, 152–161 (2010).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Huang, W., Sherman, B. & Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, W., Sherman, B. & Lempicki, R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Chen, E.Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Shabalin, A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Westra, H.-J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Bendall, S.C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Aghaeepour, N. et al. Critical assessment of automated flow cytometry data analysis techniques. Nat. Methods 10, 228–238 (2013).
Qiu, P. et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 29, 886–891 (2011).
Newell, E.W., Sigal, N., Bendall, S.C., Nolan, G.P. & Davis, M.M. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity 36, 142–152 (2012).
Amir, A.D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Shen-Orr, S.S. et al. Cell type–specific gene expression differences in complex tissues. Nat. Methods 7, 287–289 (2010).
Ahn, J. et al. DeMix: deconvolution for mixed cancer transcriptomes using raw measured data. Bioinformatics 29, 1865–1871 (2013).
Vahedi, G. et al. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol. Rev. 252, 24–40 (2013).
Ciofani, M. et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303 (2012).
Karczewski, K.J. et al. Systematic functional regulatory assessment of disease-associated variants. Proc. Natl. Acad. Sci. USA 110, 9607–9612 (2013).
Rozowsky, J. et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 7, 522 (2011).
Jee, J. et al. ACT: aggregation and correlation toolbox for analyses of genome tracks. Bioinformatics 27, 1152–1154 (2011).
Arazi, A., Pendergraft, W., Ribeiro, R., Perelson, A. & Hacohen, N. Human systems immunology: hypothesis-based modeling and unbiased data-driven approaches. Semin. Immunol. 25, 193–200 (2013).
Germain, R.N., Meier-Schellersheim, M., Nita-Lazar, A. & Fraser, I. Systems biology in immunology: a computational modeling perspective. Annu. Rev. Immunol. 29, 527–585 (2011).
Segal, E. et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 34, 166–176 (2003).
Amit, I., Regev, A. & Hacohen, N. Strategies to discover regulatory circuits of the mammalian immune system. Nat. Rev. Immunol. 11, 873–880 (2011).
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309 (2011).
Jojic, V. et al. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 14, 633–643 (2013).
Amit, I. et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 (2009).
Chevrier, N. et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell 147, 853–867 (2011).
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17 (2005).
Voineagu, I. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011).
Zhang, B. et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 153, 707–720 (2013).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
Wang, I.M. et al. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol. Syst. Biol. 8, 594 (2012).
Schadt, E.E. et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37, 710–717 (2005).
Greenawalt, D.M. et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21, 1008–1016 (2011).
Zhong, H. et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 6, e1000932 (2010).
Wishart, D.S. et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006).
Sirota, M. et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 3, 96ra77 (2011).
Dudley, J.T. et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 3, 96ra76 (2011).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Saito, R. et al. A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076 (2012).
Srivas, R. et al. Assembling global maps of cellular function through integrative analysis of physical and genetic networks. Nat. Protoc. 6, 1308–1323 (2011).
Schadt, E.E., Linderman, M., Sorenson, J., Lee, L. & Nolan, G. Computational solutions to large-scale data management and analysis. Nat. Rev. Genet. 11, 647–657 (2010).
Dudley, J.T. & Butte, A. In silico research in the era of cloud computing. Nat. Biotechnol. 28, 1181–1185 (2010).
Dudley, J.T., Pouliot, Y., Chen, R., Morgan, A.A. & Butte, A.J. Translational bioinformatics in the cloud: an affordable alternative. Genome Med. 2, 51 (2010).
Kotecha, N., Krutzik, P. & Irish, J. Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom. 53, 10.17 (2010).
Lum, P.Y. et al. Extracting insights from the shape of complex data using topology. Sci. Rep. 3, 1236 (2013).
Thomas, N., Heather, J., Ndifon, W., Shawe-Taylor, J. & Chain, B. Decombinator: a tool for fast, efficient gene assignment in T-cell receptor sequences using a finite state machine. Bioinformatics 29, 542–550 (2013).
Warren, R.L., Nelson, B. & Holt, R. Profiling model T-cell metagenomes with short reads. Bioinformatics 25, 458–464 (2009).
Barak, M., Zuckerman, N., Edelman, H., Unger, R. & Mehr, R. IgTree: creating Immunoglobulin variable region gene lineage trees. J. Immunol. Methods 338, 67–74 (2008).
Glanville, J. et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc. Natl. Acad. Sci. USA 106, 20216–20221 (2009).
Pepke, S., Wold, B. & Mortazavi, A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods 6, S22–S32 (2009).
Koboldt, D.C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Guttman, M. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28, 503–510 (2010).
Langmead, B., Schatz, M., Lin, J., Pop, M. & Salzberg, S. Searching for SNPs with cloud computing. Genome Biol. 10, R134 (2009).
Wilbanks, E.G. & Facciotti, M. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS ONE 5, e11471 (2010).
Ji, H. et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26, 1293–1300 (2008).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Rozowsky, J. et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 27, 66–75 (2009).
Kharchenko, P.V., Tolstorukov, M. & Park, P. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26, 1351–1359 (2008).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Broman, K.W., Wu, H., Sen, S. & Churchill, G. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003).
Clayton, D. & Leung, H.-T. An R package for analysis of whole-genome association studies. Hum. Hered. 64, 45–51 (2007).
Watson, M. CoXpress: differential co-expression in gene expression data. BMC Bioinformatics 7, 509 (2006).
Bonneau, R. et al. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 7, R36 (2006).
Basso, K. et al. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 37, 382–390 (2005).
Aghaeepour, N., Nikolic, R., Hoos, H. & Brinkman, R. Rapid cell population identification in flow cytometry data. Cytometry 79A, 6–13 (2011).
Pyne, S. et al. Automated high-dimensional flow cytometric data analysis. Proc. Natl. Acad. Sci. USA 106, 8519–8524 (2009).
Qian, Y. et al. Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry Clin. Cytom. 78V (suppl. 1), S69–S82 (2010).
Zare, H., Shooshtari, P., Gupta, A. & Brinkman, R. Data reduction for spectral clustering to analyze high throughput flow cytometry data. BMC Bioinformatics 11, 403 (2010).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Lefranc, M.P. et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 27, 209–212 (1999).
Siebert, J.C., Munsil, W., Rosenberg-Hasson, Y., Davis, M. & Maecker, H. The Stanford Data Miner: a novel approach for integrating and exploring heterogeneous immunological data. J. Transl. Med. 10, 62 (2012).
Pritchard, J.K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Kumar, S., Sanderford, M., Gray, V., Ye, J. & Liu, L. Evolutionary diagnosis method for variants in personal exomes. Nat. Methods 9, 855–856 (2012).
Li, M.J. et al. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 40, D1047–D1054 (2012).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Boyle, A.P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Raychaudhuri, S. et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 5, e1000534 (2009).
Griffith, M. et al. DGIdb: mining the druggable genome. Nat. Methods 10, 1209–1210 (2013).
Engreitz, J.M. et al. ProfileChaser: searching microarray repositories based on genome-wide patterns of differential expression. Bioinformatics 27, 3317–3318 (2011).
Heiser, L.M. et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. USA 109, 2724–2729 (2012).
Tan, C.M., Chen, E., Dannenfelser, R., Clark, N. & Ma'ayan, A. Network2Canvas: network visualization on a canvas with enrichment analysis. Bioinformatics 29, 1872–1878 (2013).
Acknowledgements
We thank C. Berin, B. Brown, R. Kosoy, B. Readhead and C. Tato for critical reading and feedback on the manuscript. This work was supported by funding from the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK098242) and the Pharmaceutical Research and Manufacturers of America Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kidd, B., Peters, L., Schadt, E. et al. Unifying immunology with informatics and multiscale biology. Nat Immunol 15, 118–127 (2014). https://doi.org/10.1038/ni.2787
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2787