Abstract
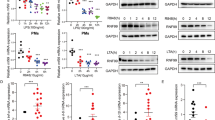
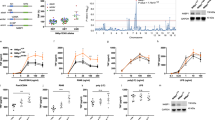
Tolerance to endotoxins that is triggered by prior exposure to Toll-like receptor (TLR) ligands provides a mechanism with which to dampen inflammatory cytokines. The receptor-interacting protein RIP140 interacts with the transcription factor NF-κB to regulate the expression of genes encoding proinflammatory cytokines. Here we found lipopolysaccharide stimulation of kinase Syk–mediated tyrosine phosphorylation of RIP140 and interaction of the NF-κB subunit RelA with RIP140. These events resulted in more recruitment of the E3 ligase SCF to tyrosine-phosphorylated RIP140, which degraded RIP140 to inactivate genes encoding inflammatory cytokines. Macrophages expressing nondegradable RIP140 were resistant to the establishment of endotoxin tolerance for specific 'tolerizable' genes. Our results identify RelA as an adaptor with which SCF fine tunes NF-κB target genes by targeting the coactivator RIP140 and show an unexpected role for RIP140 degradation in resolving inflammation and endotoxin tolerance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
O'Neill, L.A. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity 29, 12–20 (2008).
Baker, R.G., Hayden, M.S. & Ghosh, S. NF-κB, inflammation, and metabolic disease. Cell Metab. 13, 11–22 (2011).
Wen, H. et al. Fatty acid–induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 12, 408–415 (2011).
Tabas, I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 10, 36–46 (2010).
Sheedy, F.J. et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147 (2010).
Mansell, A. et al. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat. Immunol. 7, 148–155 (2006).
Foster, S.L., Hargreaves, D.C. & Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007).
Chen, J. & Ivashkiv, L.B. IFN-γ abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc. Natl. Acad. Sci. USA 107, 19438–19443 (2010).
Nathan, C. & Ding, A. Nonresolving inflammation. Cell 140, 871–882 (2010).
Biswas, S.K. & Lopez-Collazo, E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 (2009).
Park, S.H., Park-Min, K.H., Chen, J., Hu, X. & Ivashkiv, L.B. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat. Immunol. 12, 607–615 (2011).
Chan, C., Li, L., McCall, C.E. & Yoza, B.K. Endotoxin tolerance disrupts chromatin remodeling and NF-κB transactivation at the IL-1β promoter. J. Immunol. 175, 461–468 (2005).
White, R. et al. Role of RIP140 in metabolic tissues: connections to disease. FEBS Lett. 582, 39–45 (2008).
Fritah, A., Christian, M. & Parker, M.G. The metabolic coregulator RIP140: an update. Am. J. Physiol. Endocrinol. Metab. 299, E335–E340 (2010).
Lee, C.H., Chinpaisal, C. & Wei, L.N. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol. Cell. Biol. 18, 6745–6755 (1998).
Leonardsson, G. et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA 101, 8437–8442 (2004).
Christian, M. et al. RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 25, 9383–9391 (2005).
Ho, P.C., Lin, Y.W., Tsui, Y.C., Gupta, P. & Wei, L.N. A negative regulatory pathway of GLUT4 trafficking in adipocyte: new function of RIP140 in the cytoplasm via AS160. Cell Metab. 10, 516–523 (2009).
Ho, P.C., Chuang, Y.S., Hung, C.H. & Wei, L.N. Cytoplasmic receptor-interacting protein 140 (RIP140) interacts with perilipin to regulate lipolysis. Cell. Signal. 23, 1396–1403 (2011).
Ho, P.C. & Wei, L.N. Negative regulation of adiponectin secretion by receptor interacting protein 140 (RIP140). Cell Signal. 24, 71–76 (2012).
Mostaqul Huq, M.D., Gupta, P. & Wei, L.N. Post-translational modifications of nuclear co-repressor RIP140: a therapeutic target for metabolic diseases. Curr. Med. Chem. 15, 386–392 (2008).
Huq, M.D., Ha, S.G., Barcelona, H. & Wei, L.N. Lysine methylation of nuclear co-repressor receptor interacting protein 140. J. Proteome Res. 8, 1156–1167 (2009).
Gupta, P. et al. PKCɛ stimulated arginine methylation of RIP140 for its nuclear-cytoplasmic export in adipocyte differentiation. PLoS ONE 3, e2658 (2008).
Ho, P.C. et al. Modulation of lysine acetylation-stimulated repressive activity by Erk2-mediated phosphorylation of RIP140 in adipocyte differentiation. Cell. Signal. 20, 1911–1919 (2008).
Zschiedrich, I. et al. Coactivator function of RIP140 for NFκB/RelA-dependent cytokine gene expression. Blood 112, 264–276 (2008).
Ho, P.C., Chang, K.C., Chuang, Y.S. & Wei, L.N. Cholesterol regulation of receptor-interacting protein 140 via microRNA-33 in inflammatory cytokine production. FASEB J. 25, 1758–1766 (2011).
Clague, M.J. & Urbe, S. Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 (2010).
Natoli, G. & Chiocca, S. Nuclear ubiquitin ligases, NF-κB degradation, and the control of inflammation. Sci. Signal. 1, pe1 (2008).
Maine, G.N., Mao, X., Komarck, C.M. & Burstein, E. COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J. 26, 436–447 (2007).
Freudenberg, M.A. & Galanos, C. Induction of tolerance to lipopolysaccharide (LPS)-D-galactosamine lethality by pretreatment with LPS is mediated by macrophages. Infect. Immun. 56, 1352–1357 (1988).
Vats, D. et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35 (2003).
Mosser, D.M. & Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969 (2008).
Kinjyo, I. et al. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 17, 583–591 (2002).
Nakagawa, R. et al. SOCS-1 participates in negative regulation of LPS responses. Immunity 17, 677–687 (2002).
Dimitriou, I.D. et al. Putting out the fire: coordinated suppression of the innate and adaptive immune systems by SOCS1 and SOCS3 proteins. Immunol. Rev. 224, 265–283 (2008).
Hamerman, J.A., Tchao, N.K., Lowell, C.A. & Lanier, L.L. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat. Immunol. 6, 579–586 (2005).
Han, C. et al. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat. Immunol. 11, 734–742 (2010).
Hu, X., Chakravarty, S.D. & Ivashkiv, L.B. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol. Rev. 226, 41–56 (2008).
Medzhitov, R. & Horng, T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9, 692–703 (2009).
Smale, S.T. Selective transcription in response to an inflammatory stimulus. Cell 140, 833–844 (2010).
Strebovsky, J., Walker, P., Lang, R. & Dalpke, A.H. Suppressor of cytokine signaling 1 (SOCS1) limits NFκB signaling by decreasing p65 stability within the cell nucleus. FASEB J. 25, 863–874 (2011).
Yoon, C.H., Miah, M.A., Kim, K.P. & Bae, Y.S. New Cdc2 Tyr 4 phosphorylation by dsRNA-activated protein kinase triggers Cdc2 polyubiquitination and G2 arrest under genotoxic stresses. EMBO Rep. 11, 393–399 (2010).
Yoo, Y., Ho, H.J., Wang, C. & Guan, J.L. Tyrosine phosphorylation of cofilin at Y68 by v-Src leads to its degradation through ubiquitin-proteasome pathway. Oncogene 29, 263–272 (2010).
Oeckinghaus, A., Hayden, M.S. & Ghosh, S. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 (2011).
Wei, L.N., Hu, X., Chandra, D., Seto, E. & Farooqui, M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J. Biol. Chem. 275, 40782–40787 (2000).
Vo, N., Fjeld, C. & Goodman, R.H. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell. Biol. 21, 6181–6188 (2001).
Rao, M.K. & Wilkinson, M.F. Tissue-specific and cell type-specific RNA interference in vivo. Nat. Protoc. 1, 1494–1501 (2006).
Acknowledgements
We thank P.-T. Liu, S.C. Chan, K.-C. Chang, Y.-S. Chuang, A. Smith, C. Korteum and P. Lam for technical help and S. Kaech for critical reading. Supported by US National Insitutes of Health (DK60521, DK54733, DA11190 and K02-DA13926), the Distinguished McKnigh Professorship of the University of of Minnesota (L.-N. Wei) and the American Heart Association (P.-C. Ho).
Author information
Authors and Affiliations
Contributions
P.-C.H. designed and did most experiments and wrote the manuscript; Y.-C.T. did animal experiments and analyzed the results; X.F. did animal experiments; D.R.G. provided reagents; and L.-N.W. supervised and analyzed the research, wrote the manuscript and provided financial support.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–20 (PDF 3612 kb)
Rights and permissions
About this article
Cite this article
Ho, PC., Tsui, YC., Feng, X. et al. NF-κB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat Immunol 13, 379–386 (2012). https://doi.org/10.1038/ni.2238
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2238
This article is cited by
-
RNA-Seq based transcriptome analysis in oral lichen planus
Hereditas (2021)
-
Regulation of exosome secretion by cellular retinoic acid binding protein 1 contributes to systemic anti-inflammation
Cell Communication and Signaling (2021)
-
Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing
Nature Immunology (2019)
-
Glyburide and retinoic acid synergize to promote wound healing by anti-inflammation and RIP140 degradation
Scientific Reports (2018)
-
Complex regulation of LCoR signaling in breast cancer cells
Oncogene (2017)