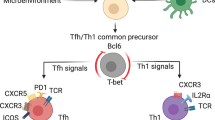
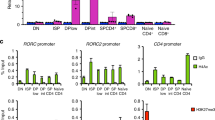
Abstract
How naive CD4+ T cells commit to the T helper type 2 (TH2) lineage is poorly understood. Here we show that the basic helix-loop-helix transcription factor Dec2 was selectively expressed in TH2 cells. CD4+ T cells from Dec2-deficient mice showed defective TH2 differentiation in vitro and in vivo in an asthma model and in response to challenge with a parasite antigen. Dec2 promoted expression of interleukin 4 (IL-4), IL-5 and IL-13 during early TH2 differentiation and directly bound to and activated transcription of genes encoding the transcription factors JunB and GATA-3. As GATA-3 induces Dec2 expression, our findings also indicate a feed-forward regulatory circuit during TH2 differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
Change history
03 September 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Dong, C. Diversification of T-helper-cell lineages: finding the family root of IL-17-producing cells. Nat. Rev. Immunol. 6, 329–333 (2006).
Glimcher, L.H. & Murphy, K.M. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 14, 1693–1711 (2000).
Martinez, G.J., Nurieva, R.I., Yang, X.O. & Dong, C. Regulation and function of proinflammatory TH17 cells. Ann. NY Acad. Sci. 1143, 188–211 (2008).
Dong, C. Regulation and pro-inflammatory function of interleukin-17 family cytokines. Immunol. Rev. 226, 80–86 (2008).
Dong, C. & Flavell, R.A. Cell fate decision: T-helper 1 and 2 subsets in immune responses. Arthritis Res. 2, 179–188 (2000).
Lederer, J.A. et al. Cytokine transcriptional events during helper T cell subset differentiation. J. Exp. Med. 184, 397–406 (1996).
Angkasekwinai, P. et al. Interleukin 25 promotes the initiation of proallergic type 2 responses. J. Exp. Med. 204, 1509–1517 (2007).
Nurieva, R.I. et al. A costimulation-initiated signaling pathway regulates NFATc1 transcription in T lymphocytes. J. Immunol. 179, 1096–1103 (2007).
Dong, C., Nurieva, R.I. & Prasad, D.V. Immune regulation by novel costimulatory molecules. Immunol. Res. 28, 39–48 (2003).
Cote-Sierra, J. et al. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. USA 101, 3880–3885 (2004).
Yamane, H., Zhu, J. & Paul, W.E. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J. Exp. Med. 202, 793–804 (2005).
Liao, W. et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor α-chain expression. Nat. Immunol. 9, 1288–1296 (2008).
Azmi, S. & Taneja, R. Embryonic expression of mSharp-1/mDEC2, which encodes a basic helix-loop-helix transcription factor. Mech. Dev. 114, 181–185 (2002).
Fujimoto, K. et al. Molecular cloning and characterization of DEC2, a new member of basic helix-loop-helix proteins. Biochem. Biophys. Res. Commun. 280, 164–171 (2001).
Honma, S. et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419, 841–844 (2002).
Azmi, S., Ozog, A. & Taneja, R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J. Biol. Chem. 279, 52643–52652 (2004).
Sato, F. et al. Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates vascular endothelial growth factor expression. Genes Cells 13, 131–144 (2008).
Li, B., Tournier, C., Davis, R.J. & Flavell, R.A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18, 420–432 (1999).
Hartenstein, B. et al. Th2 cell-specific cytokine expression and allergen-induced airway inflammation depend on JunB. EMBO J. 21, 6321–6329 (2002).
Bunting, K., Wang, J. & Shannon, M.F. Control of interleukin-2 gene transcription: a paradigm for inducible, tissue-specific gene expression. Vitam. Horm. 74, 105–145 (2006).
Jain, J., Loh, C. & Rao, A. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 7, 333–342 (1995).
Jain, J., Valge-Archer, V.E. & Rao, A. Analysis of the AP-1 sites in the IL-2 promoter. J. Immunol. 148, 1240–1250 (1992).
Mohrs, M., Shinkai, K., Mohrs, K. & Locksley, R.M. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity 15, 303–311 (2001).
Zhu, J. et al. Conditional deletion of Gata3 shows its essential function in TH1-TH2 responses. Nat. Immunol. 5, 1157–1165 (2004).
Pearce, E.J., Kane, C.M., Sun, J., Taylor, J.J., McKee, A.S. & Cervi, L. Th2 response polarization during infection with the helminth parasite Schistosoma mansoni. Immunol. Rev. 201, 117–126 (2004).
Yang, X.O. et al. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 205, 1063–1075 (2008).
Frazer, K.A., Pachter, L., Poliakov, A., Rubin, E.M. & Dubchak, I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–279 (2004).
Mayor, C. et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 (2000).
Farre, D. et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31, 3651–3653 (2003).
Messeguer, X. et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 (2002).
Zhumabekov, T., Corbella, P., Tolaini, M. & Kioussis, D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J. Immunol. Methods 185, 133–140 (1995).
Dong, C. & Flavell, R.A. Control of T helper cell differentiation–in search of master genes. Sci. STKE 2000, PE1 (2000).
Nurieva, R. et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483 (2007).
Yang, X.O. et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282, 9358–9363 (2007).
Boros, D.L. & Warren, K.S. Delayed hypersensitivity-type granuloma formation and dermal reaction induced and elicited by a soluble factor isolated from Schistosoma mansoni eggs. J. Exp. Med. 132, 488–507 (1970).
Ouyang, W. et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity 9, 745–755 (1998).
Yang, X.O. et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity 28, 29–39 (2008).
Voice, J. et al. c-Maf and JunB mediation of Th2 differentiation induced by the type 2 G protein-coupled receptor (VPAC2) for vasoactive intestinal peptide. J. Immunol. 172, 7289–7296 (2004).
Evans, K.E. & Fox, S.W. Interleukin-10 inhibits osteoclastogenesis by reducing NFATc1 expression and preventing its translocation to the nucleus. BMC Cell Biol. 8, 4 (2007).
Akimzhanov, A.M., Yang, X.O. & Dong, C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 282, 5969–5972 (2007).
Acknowledgements
We thank S. Rivera and J. Parker-Thornburg of the MD Anderson Genetic Engineered Mouse Facility for help in generating knockout and transgenic animals; K. Murphy (Washington University) for the GFP-RV and GFP-RV-GATA-3 vectors; Y. Belkaid (National Institute of Allergy and Infectious Diseases, National Institutes of Health) for S. mansoni eggs and suggestions; and members of the Dong laboratory for help and discussion. Supported by the National Institutes of Health (C.D. and R.N.), the MD Anderson Center for Targeted Therapy (C.D. and R.N.), the American Heart Association (X.O.Y. and R.N.), the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (J.Z.), the Ministry of Education of China (J.P.) and the Leukemia and Lymphoma Society (C.D.).
Author information
Authors and Affiliations
Contributions
C.D., X.O.Y. and B.S. designed the research, analyzed and interpreted the results; X.O.Y., P.A., J.Z., J.P., Z.L., R.N., X.L., Y.C. and S.H.C. did the experiments; and X.O.Y., B.S. and C.D. prepared the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 358 kb)
Rights and permissions
About this article
Cite this article
Yang, X., Angkasekwinai, P., Zhu, J. et al. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat Immunol 10, 1260–1266 (2009). https://doi.org/10.1038/ni.1821
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1821
This article is cited by
-
Single cell RNA-sequencing identified Dec2 as a suppressive factor for spermatogonial differentiation by inhibiting Sohlh1 expression
Scientific Reports (2019)
-
Loss of the basic helix-loop-helix transcription factor Bhlhe41 induces cell death and impairs neurite outgrowth in Neuro2a cells
Molecular and Cellular Biochemistry (2019)
-
The basic helix-loop-helix transcription factor SHARP1 is an oncogenic driver in MLL-AF6 acute myelogenous leukemia
Nature Communications (2018)
-
Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells
Nature Immunology (2017)
-
Regulation of B-1a cells: another novel function of the basic helix-loop-helix transcriptional regulator BHLHE41
Cellular & Molecular Immunology (2017)