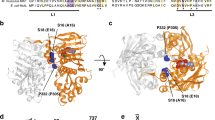
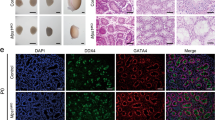
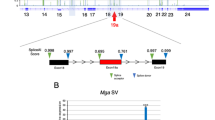
Abstract
MutL homolog 3 (Mlh3) is a member of a family of proteins conserved during evolution and having dual roles in DNA mismatch repair and meiosis1,2. The pathway in eukaryotes consists of the DNA-binding components, which are the homologs of the bacterial MutS protein (MSH 2–6), and the MutL homologs, which bind to the MutS homologs and are essential for the repair process. Three of the six homologs of MutS that function in these processes, Msh2, Msh3 and Msh6, are involved in the mismatch repair of mutations, frameshifts and replication errors2,3, and two others, Msh4 and Msh5, have specific roles in meiosis4,5,6,7,8,9,10. Of the four MutL homologs, Mlh1, Mlh3, Pms1 and Pms2, three are involved in mismatch repair and at least two, Pms2 and Mlh1, are essential for meiotic progression in both yeast and mice2,3,11,12,13,14. To assess the role of Mlh3 in mammalian meiosis, we have generated and characterized Mlh3−/− mice15. Here we show that Mlh3−/− mice are viable but sterile. Mlh3 is required for Mlh1 binding to meiotic chromosomes and localizes to meiotic chromosomes from the mid pachynema stage of prophase I. Mlh3−/− spermatocytes reach metaphase before succumbing to apoptosis, but oocytes fail to complete meiosis I after fertilization. Our results show that Mlh3 has an essential and distinct role in mammalian meiosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Modrich, P. & Lahue, R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65, 101–133 (1996).
Kolodner, R.D. & Marsischky, G.T. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9, 89–96 (1999).
Wang, T.F., Kleckner, N. & Hunter, N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl Acad. Sci. USA 96, 13914–13919 (1999).
Hollingsworth, N.M., Ponte, L. & Halsey, C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9, 1728–1739 (1995).
Ross-Macdonald, P. & Roeder, G.S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79, 1069–1080 (1994).
Her, C., Wu, X., Wan, W. & Doggett, N.A. Identification and characterization of the mouse MutS homolog 5: Msh5. Mamm. Genome 10, 1054–1061 (1999).
de Vries, S.S. et al. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13, 523–531 (1999).
Kneitz, B. et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097 (2000).
Paquis-Flucklinger, V. et al. Cloning and expression analysis of a meiosis-specific MutS homolog: the human MSH4 gene. Genomics 44, 188–194 (1997).
Edelmann, W. et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nature Genet. 21, 123–127 (1999).
Hunter, N. & Borts, R.H. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 11, 1573–1582 (1997).
Baker, S.M. et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Genet. 13, 336–342 (1996).
Baker, S.M. et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 82, 309–319 (1995).
Edelmann, W. et al. Meiotic pachytene arrest in MLH-1-deficient mice. Cell 85, 1125–1134 (1996).
Lipkin, S.M. et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nature Genet. 24, 27–35 (2000).
Moens, P.B. et al. The time course and localization of recombination-related proteins at meiosis in the mouse conform to models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611–1622 (2002).
Anderson, L.K., Reeves, A., Webb, L.M. & Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151, 1569–1579 (1999).
Flores-Rozas, H. & Kolodner, R.D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl Acad. Sci. USA 95, 12404–12409 (1998).
Woods, L.M. et al. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 145, 1395–1406 (1999).
Cohen, P.E. & Pollard, J.W. Regulation of meiotic recombination and prophase I progression in mammals. BioEssays 23, 996–1009 (2001).
Guarne, A., Junop, M.S. & Yang, W. Structure and function of the N-terminal 40 kDa fragment of human PMS2: a monomeric GHL ATPase. EMBO J. 20, 5521–5531 (2001).
Osoegawa, K. et al. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10, 116–128 (2000).
Marra, G. et al. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc. Natl Acad. Sci. USA 95, 8568–8573 (1998).
Green, E.D. Strategies for the systematic sequencing of complex genomes. Nature Rev. Genet. 2, 573–583 (2001).
Ellsworth, R.E. et al. Comparative genomic sequence analysis of the human and mouse cystic fibrosis transmembrane conductance regulator genes. Proc. Natl Acad. Sci. USA 97, 1172–1177 (2000).
Touchman, J.W. et al. The genomic region encompassing the nephropathic cystinosis gene (CTNS): complete sequencing of a 200-kb segment and discovery of a novel gene within the common cystinosis-causing deletion. Genome Res. 10, 165–173 (2000).
Ewing, B., Hillier, L., Wendl, M.C. & Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185 (1998).
Edelmann, W. et al. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res. 60, 803–807 (2000).
Wang, D. & Enders, G.C. Expression of a specific mouse germ cell nuclear antigen (GCNA1) by early embryonic testicular teratoma cells in 129/Sv-Sl/+ mice. Cancer Lett. 100, 31–36 (1996).
Acknowledgements
We thank G. Elliott and A. Becker for help with mouse studies; M. Liskay and W. Edelmann for critically reading this manuscript; G. Enders for antibodies; and N. Kolas for discussions and advice. This work was supported by the Albert Einstein College of Medicine (P.E.C.), National Human Genome Research Institute intramural funds and New Faculty start-up funds through the University of California, Irvine (S.M.L.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Lipkin, S., Moens, P., Wang, V. et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 31, 385–390 (2002). https://doi.org/10.1038/ng931
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng931
This article is cited by
-
High amount of fertility reducing tumors and procedures, but no evidence for premature ovarian failure in female Lynch syndrome patients
Familial Cancer (2024)
-
DNA double-strand break genetic variants in patients with premature ovarian insufficiency
Journal of Ovarian Research (2023)
-
A novel recombination protein C12ORF40/REDIC1 is required for meiotic crossover formation
Cell Discovery (2023)
-
Potential Involvement of DNA Methylation in Hybrid Sterility in Hermaphroditic Argopecten Scallops
Marine Biotechnology (2023)
-
Mouse oocytes carrying metacentric Robertsonian chromosomes have fewer crossover sites and higher aneuploidy rates than oocytes carrying acrocentric chromosomes alone
Scientific Reports (2022)