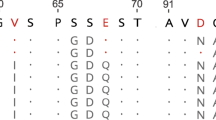
Abstract
Similar morphological or physiological changes occurring in multiple evolutionary lineages are not uncommon. Such parallel changes are believed to be adaptive, because a complex character is unlikely to originate more than once by chance. However, the occurrence of adaptive parallel amino acid substitutions is debated1,2,3. Here I propose four requirements for establishing adaptive parallel evolution at the protein sequence level and use these criteria to demonstrate such a case. I report that the gene encoding pancreatic ribonuclease was duplicated independently in Asian and African leaf-eating monkeys. Statistical analyses of DNA sequences, functional assays of reconstructed ancestral proteins and site-directed mutagenesis show that the new genes acquired enhanced digestive efficiencies through parallel amino acid replacements driven by darwinian selection. They also lost a non-digestive function independently, under a relaxed selective constraint. These results demonstrate that despite the overall stochasticity, even molecular evolution has a certain degree of repeatability and predictability under the pressures of natural selection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Doolittle, R.F. Convergent evolution: the need to be explicit. Trends Biochem. Sci. 19, 15–18 (1994).
Zhang, J. & Kumar, S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol. Biol. Evol. 14, 527–536 (1997).
Zhang, J. Parallel functional changes in the digestive RNases of ruminants and colobines by divergent amino acid substitutions. Mol. Biol. Evol. 20, 1310–1317 (2003).
Stewart, C.B., Schilling, J.W. & Wilson, A.C. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 330, 401–404 (1987).
Yokoyama, R. & Yokoyama, S. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proc. Natl. Acad. Sci. USA 87, 9315–9318 (1990).
Rodriguez-Trelles, F., Tarrio, R. & Ayala, F.J. Convergent neofunctionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene. Proc. Natl. Acad. Sci. USA 100, 13413–13417 (2003).
Mundy, N.I. A window on the genetics of evolution: MC1R and plumage colouration in birds. Proc. Biol. Sci. 272, 1633–1640 (2005).
Delsen, E. Evolutionary history of the colobine monkeys in paleoenvironmental perspective. in Colobine Monkeys: Their Ecology, Behaviour and Evolution (eds. Davies, A.G. & Oates, J.F.) 11–44 (Cambridge Univ. Press, Cambridge, UK, 1994).
Kay, R.N.B. & Davies, A.G. Digestive physiology. in Colobine Monkeys: Their Ecology, Behaviour and Evolution (eds. Davies, A.G. & Oates, J.F.) 229–249 (Cambridge Univ. Press, Cambridge, UK, 1994).
Barnard, E.A. Biological function of pancreatic ribonuclease. Nature 221, 340–344 (1969).
Beintema, J.J. The primary structure of langur (Presbytis entellus) pancreatic ribonuclease: adaptive features in digestive enzymes in mammals. Mol. Biol. Evol. 7, 470–477 (1990).
Guyton, A.C. & Hall, J.E. Textbook of Medical Physiology (Saunders, London, 1996).
Zhang, J., Zhang, Y.P. & Rosenberg, H.F. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30, 411–415 (2002).
Tang, H., Wyckoff, G.J., Lu, J. & Wu, C.I. A universal evolutionary index for amino acid changes. Mol. Biol. Evol. 21, 1548–1556 (2004).
Jermann, T.M., Opitz, J.G., Stackhouse, J. & Benner, S.A. Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature 374, 57–59 (1995).
Futami, J. et al. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 16, 413–419 (1997).
Sorrentino, S. & Libonati, M. Human pancreatic-type and nonpancreatic-type ribonucleases: a direct side-by-side comparison of their catalytic properties. Arch. Biochem. Biophys. 312, 340–348 (1994).
Sorrentino, S., Naddeo, M., Russo, A. & D'Alessio, G. Degradation of double-stranded RNA by human pancreatic ribonuclease: crucial role of noncatalytic basic amino acid residues. Biochemistry 42, 10182–10190 (2003).
Bull, J.J. et al. Exceptional convergent evolution in a virus. Genetics 147, 1497–1507 (1997).
Carroll, S.B. Chance and necessity: the evolution of morphological complexity and diversity. Nature 409, 1102–1109 (2001).
Monod, J. Chance and Necessity (Vintage, New York, 1972).
Magness, C.L. et al. Analysis of the Macaca mulatta transcriptome and the sequence divergence between Macaca and human. Genome Biol. 6, R60 (2005).
Sawyer, S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6, 526–538 (1989).
Zhang, J. & Nei, M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J. Mol. Evol. 44 (Suppl.), S139–S146 (1997).
Yang, Z., Kumar, S. & Nei, M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141, 1641–1650 (1995).
Tajima, F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135, 599–607 (1993).
Zhang, J., Rosenberg, H.F. & Nei, M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95, 3708–3713 (1998).
Zhang, J., Kumar, S. & Nei, M. Small-sample tests of episodic adaptive evolution: a case study of primate lysozymes. Mol. Biol. Evol. 14, 1335–1338 (1997).
Li, W. Molecular Evolution (Sinauer, Sunderland, Massachusetts, USA, 1997).
Zhang, J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 50, 56–68 (2000).
Acknowledgements
I thank M. Bakewell, S. Cho, W. Grus, A. Rooney and D. Webb for comments. This work was supported by the US National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary Fig. 1
The structures of the human and guereza pancreatic RNase genes. (PDF 17 kb)
Rights and permissions
About this article
Cite this article
Zhang, J. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat Genet 38, 819–823 (2006). https://doi.org/10.1038/ng1812
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1812
This article is cited by
-
Molecular convergent and parallel evolution among four high-elevation anuran species from the Tibetan region
BMC Genomics (2020)
-
Sex alters molecular evolution in diploid experimental populations of S. cerevisiae
Nature Ecology & Evolution (2020)
-
Evolution of the bitter taste receptor TAS2R38 in colobines
Primates (2020)
-
Cluster expansion of apolipoprotein D (ApoD) genes in teleost fishes
BMC Evolutionary Biology (2019)
-
Duplication and parallel evolution of the pancreatic ribonuclease gene (RNASE1) in folivorous non-colobine primates, the howler monkeys (Alouatta spp.)
Scientific Reports (2019)