Abstract
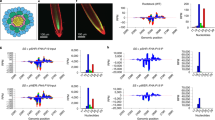
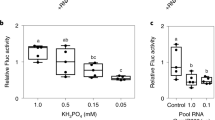
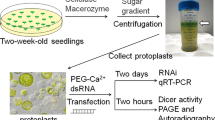
In RNA interference1,2, the RNase-III enzyme Dicer3 processes exogenous double-stranded RNA into small interfering RNAs (siRNAs). siRNAs guide RNA-induced silencing complexes to cleave homologous transcripts, enabling gene-specific knock-down4. In plants, double-stranded RNA is processed into siRNA species of 21 nucleotides (nt) and 24 nt (ref. 5), but, unlike in nematodes6, the Dicer enzymes involved in this processing have not been identified. Additionally, in both plants and nematodes, systemic signals7,8,9,10 with RNA components convey the sequence-specific effects of RNA interference between cells. Here, we describe Arabidopsis thaliana mutants with altered silencing cell-to-cell movement beyond the vasculature. At least three SILENCING MOVEMENT DEFICIENT genes (SMD1, SMD2 and SMD3) are required for trafficking, the extent of which correlates with siRNA levels in the veins. Five alleles defective in synthesis of 21-nt, but not 24-nt, siRNAs carry mutations in Dicer-like 4 (DCL4) that are involved in biogenesis of trans-acting siRNAs11,12. We show that the biogenesis and function of trans-acting siRNA can be genetically uncoupled from a bona fide DCL4-dependent pathway that accounts for RNA interference and for production of the 21-nt siRNA component of the plant cell-to-cell silencing signal.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Waterhouse, P.M. & Helliwell, C.A. Exploring plant genomes by RNA-induced gene silencing. Nat. Rev. Genet. 4, 29–38 (2003).
Bernstein, E., Caudy, A.A., Hammond, S.M. & Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
Hammond, S.M., Bernstein, E., Beach, D. & Hannon, G. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature 404, 293–296 (2000).
Hamilton, A., Voinnet, O., Chappell, L. & Baulcombe, D.C. Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 (2002).
Mello, C.C. & Conte, D. Jr. Revealing the world of RNA interference. Nature 431, 338–342 (2004).
Palauqui, J.-C., Elmayan, T., Pollien, J.-M. & Vaucheret, H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745 (1997).
Voinnet, O. & Baulcombe, D.C. Systemic signalling in gene silencing. Nature 389, 553 (1997).
Voinnet, O., Vain, P., Angell, S. & Baulcombe, D.C. Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell 95, 177–187 (1998).
Winston, W.M., Molodowitch, C. & Hunter, C.P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002).
Gasciolli, V., Mallory, A.C., Bartel, D.P. & Vaucheret, H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15, 1494–1500 (2005).
Xie, Z., Allen, E., Wilken, A. & Carrington, J.C. DICER-LIKE4 functions in trans-acting siRNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102, 12984–12989 (2005).
Tang, G., Reinhart, B.J., Bartel, D.P. & Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63 (2003).
Xie, Z. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, E104 (2004).
Bartel, B. & Bartel, D.P. MicroRNAs: at the root of plant development? Plant Physiol. 132, 709–717 (2003).
Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L. & Poethig, R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379 (2004).
Himber, C., Dunoyer, P., Moissiard, G., Ritzenthaler, C. & Voinnet, O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22, 4523–4533 (2003).
Schwach, F., Vaistij, F.E., Jones, L. & Baulcombe, D.C. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852 (2005).
Allen, E., Xie, Z., Gustafson, A.M. & Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005).
Carmell, M.A., Xuan, Z., Zhang, M.Q. & Hannon, G.J. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16, 2733–2742 (2002).
Mourrain, P. et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542 (2000).
Dalmay, T., Hamilton, A.J., Rudd, S., Angell, S. & Baulcombe, D.C. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553 (2000).
Elbashir, S.M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001).
Szittya, G., Molnar, A., Silhavy, D., Hornyik, C. & Burgyan, J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14, 359–372 (2002).
Acknowledgements
We thank P. Brodersen and L. Navarro for critical reading of the manuscript, S. MacFarlane for providing TRV-PDS, M. Alioua for technical support and R. Wagner's team for greenhouse work. Research in our laboratory is supported by the Centre National de la Recherche Scientifique and an EMBO Young Investigator award attributed to O.V.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Northern analysis of SULPHUR mRNA accumulation in the four mutant classes. (PDF 259 kb)
Supplementary Fig. 2
Quantitative RT-PCR (RT-qPCR) analyses of ta-siRNA target accumulation in the five independant alleles of class IV mutant. (PDF 916 kb)
Rights and permissions
About this article
Cite this article
Dunoyer, P., Himber, C. & Voinnet, O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37, 1356–1360 (2005). https://doi.org/10.1038/ng1675
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1675
This article is cited by
-
A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants
Plant Methods (2020)
-
Mobile RNAs—the magical elf traveling between plant and the associated organisms
ExRNA (2019)
-
Genome-wide identification of the Dicer-like family in cotton and analysis of the DCL expression modulation in response to biotic stress in two contrasting commercial cultivars
BMC Plant Biology (2019)
-
Long DCL4-substrate dsRNAs efficiently induce RNA interference in plant cells
Scientific Reports (2019)
-
A proteomic study of cysteine protease induced cell death in anthers of male sterile tobacco transgenic plants
Physiology and Molecular Biology of Plants (2019)