Abstract
Characterizing fine-scale variation in human recombination rates is important, both to deepen understanding of the recombination process1 and to aid the design of disease association studies2,3. Current genetic maps show that rates vary on a megabase scale, but studying finer-scale variation using pedigrees is difficult. Sperm-typing experiments4,5,6 have characterized regions where crossovers cluster into 1–2-kb hot spots, but technical difficulties limit the number of studies7. An alternative is to use population variation to infer fine-scale characteristics of the recombination process. Several surveys8,9,10 reported 'block-like' patterns of diversity, which may reflect fine-scale recombination rate variation11,12,13, but limitations of available methods made this impossible to assess. Here, we applied a new statistical method, which overcomes these limitations, to infer patterns of fine-scale recombination rate variation in 74 genes. We found extensive rate variation both within and among genes. In particular, recombination hot spots are a common feature of the human genome: 47% (35 of 74) of genes showed substantive evidence for a hot spot, and many more showed evidence for some rate variation. No primary sequence characteristics are consistently associated with precise hot-spot location, although G+C content and nucleotide diversity are correlated with local recombination rate.
Similar content being viewed by others
Main
Population-level variation carries important information about fine-scale recombination rates14. Crossover events tend to reduce population-level association (commonly known as linkage disequilibrium; LD) among alleles on either side of the crossover, and so the expected amount of LD between sites depends on the recombination rate between them2. Until now, however, methods used to extract this information, such as plots of pair-wise LD measures, did not distinguish reliably between patterns that are due to recombination rate variation and patterns that could arise by chance, with no rate variation11,12. We used a statistical method that can make this distinction and can reliably predict the presence of recombination hot spots (defined as a small region where crossovers occur more than ten times more frequently than in surrounding sequence) from population data15.
To briefly describe the method: assume haplotypes are available or have been estimated. The method considers each sampled haplotype in turn and attempts to construct it as a mosaic of previously considered haplotypes. Under simplifying assumptions about population demography, the method uses the average length of mosaic pieces to estimate the local background recombination rate, and the positions of breaks in the mosaic to estimate the location and intensity of hot spots. It also measures the strength of the evidence for a hot spot, using a Bayes factor (BF), which is the probability of obtaining the data if a hot spot is present divided by the probability of obtaining the data if a hot spot is not present. The BF is akin to a likelihood ratio, but it integrates over (rather than maximizing over) the uncertainty in haplotypic phase and putative intensity and location of a hot spot. A BF of 10 indicates that the data are ten times more likely to be obtained if there is a hot spot than if there is not, and we consider this substantive evidence for a hot spot. In computing the BF, the method takes into account chance clustering of recombination events, or recombinant haplotypes drifting to high frequency, thus guarding against overconfidence that 'block-like' patterns of LD are due to recombination rate variation.
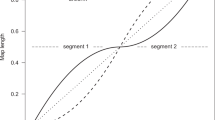
To illustrate the approach, we applied it to previously published genotype data from 50 individuals from the United Kingdom of European descent at five regions of the MHC where sperm typing identified recombination hot spots5. Polymorphism-based methods and sperm-typing methods measure different things. Sperm typing measures meiotic recombination rates in a few extant males, whereas patterns of polymorphism are shaped by recombination in ancestors of sampled individuals and, therefore, reflect average recombination rates in transmissions from many males and females over thousands of generations. Differences between results from the two approaches might therefore point to such biological mechanisms as selection or sex-specific fine-scale recombination. For these data, however, results from the two approaches agree closely: four regions showed substantive evidence in the polymorphism data for a hot spot (BF = 35, >100, >100, >100), and the fifth, suggested by sperm-typing results to have an intensity close to our threshold of ten times the background rate, showed mild evidence (BF ≈ 1.3). The estimated locations and intensities of hot spots from the polymorphism data (Fig. 1) were consistent with sperm-typing results, although we limited intensities to <100, which hindered direct comparison of estimated intensities.
The dotted, dashed and dotted-dashed green lines show estimates from three independent applications of our method; the thicker solid line shows their average. In each interval between consecutive SNPs, the plot shows the posterior mean of the intensity in that interval, relative to the background rate of recombination. The step-like appearance of the plots therefore reflects uncertainty in the estimated location of the hot spot. The vertical black lines show the approximate hot-spot center and the vertical dotted gray lines show the approximate boundaries of the hot spots, both estimated from sperm-typing results5.
We applied the same method to search for hot spots in 74 candidate genes (Supplementary Table 1 online) that we resequenced in 23 European Americans and 24 African Americans. Although previous simulation results15 suggested that the method is relatively robust to population structure, we minimized potential effects by analyzing the two population samples separately. Thirty-five genes (47%) showed substantive evidence for a hot spot in one or both samples: 19 in both samples, 11 in African Americans only and 5 in European Americans only (Fig. 2a). Estimated intensities in these genes ranged from 20 to 80, although at least two-thirds had results suggestive of intensities >100. Even with our conservative limit on hot-spot intensity and assumption of no more than one hot spot per gene, ∼60% of recombinations in these 35 genes occurred in hot spots, which covered ∼6% of the sequence. Most hot spots were narrow (only two spanned >4 kb), but our method assumed that most hot spots would be narrow, and hot-spot boundaries are difficult to estimate precisely from polymorphism data. Indeed, the notion of a hard boundary may itself be misplaced5. There was no obvious pattern of hot-spot location within a gene: estimated hot-spot centers lay in introns, exons and upstream regions.
Colored background shading indicates regions of the graph that correspond to genes with substantive evidence for a hot spot (BF > 10) in one or both samples. All axes are on a log scale. (a) Results for real data: x axis, BF for the African American (AA) sample; y axis, BF for the European American (EA) sample. (b,c) Results for simulated data without a hot spot (b) and with a hot spot (c). The gene name indicates which gene the simulated data matched for SNP density, length and background levels of recombination. (b) The color of the gene name indicates the assumed ratio of rate of gene conversion to crossing-over (black, 0; magenta, 1; orange, 10). (c) The color of the gene name indicates the intensity of the simulated hot spot: black, 1–10 (i.e., no hot spot); red, 10–20; green, 20–50; blue, 50–100.
Figure 3 shows the 90% credible intervals (CIs) for estimated hot-spot intensities. Several genes without substantive evidence for a hot spot nevertheless show evidence for some rate variation (CI excluding 1). Others have CIs that cover almost the entire range of possible intensities, suggesting that their polymorphism data are uninformative. Few genes show strong evidence against a hot spot (Figs. 2a and 3): no genes have a BF <0.1 in either population, and only eight have CIs that exclude intensities >10. On this basis, ∼90% of genes could contain hot spots.
Genes are listed, from top to bottom, in order of decreasing BF in the African American sample. Dark shading indicates those genes with substantive evidence for a hot spot (BF > 10) in Figure 2a. The vertical black line in each box indicates the median of the posterior distribution of intensity. The x axis is on a log scale.
One potential concern is that deviations from the method's simplistic modeling assumptions could lead to detection of hot spots where none exist. To test this, we applied the method to data sets simulated with constant recombination rate and varying levels of gene conversion under a demographic scenario in which one ancestral population splits into two populations, one of which undergoes a bottleneck (mimicking a European American population) and the other of which undergoes an expansion (mimicking an African American population), with limited recent migration from European Americans to African Americans (mimicking admixture). We simulated 74 data sets to match actual genes and population samples. In contrast to the real data, only one simulated data set showed substantive evidence for a hot spot in either population, and many showed substantive evidence against a hot spot (Fig. 2b). Further simulations under different assumptions showed similar patterns (Supplementary Fig. 1 online). These simulations represent some of the most plausible deviations from the modeling assumptions underlying the method2 and include scenarios previously shown to have the largest effect15,16.
One factor not included in our modeling assumptions or simulations is selection. Unlike population demography, however, selection will influence each locus differently, and so is less likely to produce large-scale, systematic distortion of the results. Furthermore, we found no relationship between genes identified here as containing hot spots and those whose polymorphism data suggest strong selective pressure (J. Akey, personal communication), and genes with particular molecular functions were no more likely to be identified as containing a hot spot (Supplementary Table 2 online). Thus, although we considered genes from specific functional pathways, our results should be representative of genes across the genome.
The large number of hot spots identified in our data seems not to be an artifact caused by deviations from simplistic modeling assumptions, but rather reflects a genuinely high frequency of recombination hot spots throughout the human genome. If we assume that the 35 genes with substantive evidence for a hot spot have a single hot spot, and no other genes have hot spots, then the hot-spot frequency is one per 63 kb. This estimate seems conservative, as it ignores the possibility of multiple hot spots in a gene5 or of genes containing hot spots but having inconclusive LD data. Thus, in human genes, hot spots are probably at least as frequent as in the human MHC class genes (one every 60–90 kb) and may be as frequent as in Saccharomyces cerevisiae (one every 50 kb)17.
We identified several genes (11 in African Americans and 5 in European Americans) that show substantive evidence for a hot spot in one population but not the other, which raises the question of whether populations differ in their recombination landscape. Because the recombination process varies among individuals18,19 and through evolutionary time20,21, differences in hot-spot characteristics across populations are not implausible. But patterns of LD reflect the average recombination process in the ancestors of a population, and not the average over extant individuals. Because human populations share much of their evolutionary history, even large differences in the recombination landscape of extant populations may be difficult to detect from LD data.
Nonetheless, we assessed support for population differences in three ways. First we examined, using simulations, how often we should expect a BF >10 in only one population sample if the populations share common hot-spot characteristics. Results (Fig. 2c) suggest that this is not unusual for the sample sizes, densities of single-nucleotide polymorphisms (SNPs) and levels of population differentiation in our data. Second, for genes showing substantive evidence for a hot spot in only one population sample, we carried out a likelihood ratio (LR) test of the hypothesis that there is a hot spot of the same intensity in both populations. The most significant difference was in CSF3R (LR = 11.3, P = 0.03). Finally, we visually inspected estimated hot-spot locations in genes showing substantive evidence for a hot spot (Supplementary Fig. 2 online) to identify genes where estimated hot-spot locations differed between samples. Although most genes had similar estimated hot-spot locations across samples (e.g., Fig. 4a and Supplementary Fig. 2 online), a few did not, including PLAUR (Fig. 4b) and CD36 (Fig. 4c). Analysis with a two-hot-spot model, however, suggested that PLAUR has two hot spots in both populations and that the intensity of the left-hand hot spot might differ between populations (LR = 9.2, P = 0.04). A similar analysis of CD36 suggested that it might also have two hot spots and that the intensity of the right-hand hot spot might differ between populations (LR = 12.7; P = 0.02). These LR and P values must be interpreted carefully (see Supplementary Methods online), but they provide a useful guide to the evidence for differences among populations and suggest genes for further investigation by sperm typing.
See legend to Figure 1 for details on interpretation. Estimated locations for the two population samples are similar in many genes, including SELP (a). Exceptions include PLAUR (b) and CD36 (c). See Supplementary Figure 2 online for similar plots for other genes.
Our method also estimates a background recombination rate for each gene, and these varied substantially among genes (Supplementary Table 3 online). As expected, estimates from the two population samples were highly correlated (correlation = 0.79). Estimates for European Americans were typically substantially lower than for African Americans (median ratio = 0.29), emphasizing the tendency for LD to extend over greater distances in European Americans. Consistent with previous reports22,23, nucleotide diversity (African Americans, P = 0.001; European Americans, P = 0.0005) and pedigree-based recombination rate estimates (African Americans, P < 0.0001; European Americans, P < 0.00005) were positively correlated with estimated local background recombination rates, as was G+C content (African Americans, P < 0.0001; European Americans, P = 0.02; Supplementary Tables 1 and 3 online). In contrast, there was no relationship between recombination rate and frequency of interspersed repeat elements (African Americans, P = 0.59; European Americans, P = 0.75). When we used multiple linear regression to control for pedigree-based estimates, the effects of both G+C content and nucleotide diversity remained significant in African Americans, and diversity remained significant in European Americans (Table 1). This suggests that the effects of nucleotide diversity and G+C content operate on a finer scale than the megabase scale on which pedigree-based estimates are obtained. (The generally larger P values in European Americans may reflect lower precision of estimates due to lower SNP density; Supplementary Tables 1 and 3 online.) Although G+C content has been correlated with recombination rates in humans on a megabase scale18,24, this is the first observation to our knowledge of finer-scale correlation.
G+C content and diversity have been previously associated with the existence of hot spots: high G+C content is associated with presence of hot spots in S. cerevisiae25, and the TAP2 (ref. 21) and β-globin26 hot spots in humans coincide with regions of higher nucleotide diversity. But we found no evidence for either effect in our data. There were no differences in G+C content of genes with substantive evidence for a hot spot versus genes without (t-test: African Americans, P = 0.34; European Americans, P = 0.36), and for genes with substantive evidence for a hot spot, there was no significant difference in the G+C content inside versus outside of estimated hot-spot boundaries (boot-strap: African Americans, P = 0.45; European Americans, P = 0.46). Similarly, we found no evidence for increased diversity in hot spots (paired t-test based on nucleotide diversity: African Americans, P = 0.32; European Americans, P = 0.75; paired t-test based on SNP density: African Americans, P = 0.06; European Americans, P = 0.65). This suggests that, in humans, although G+C content and diversity may influence the background recombination rate on a scale of tens of kilobases, any effect they have on the existence of narrow hot spots is subtle or inconsistent across genes.
We searched genes for DNA sequence elements previously hypothesized to influence hot-spot existence. None was consistently associated with the presence of hot spots (Supplementary Table 4 online), supporting previous suggestions that any relationship between primary sequence and hot-spot location is complex1,25. Although the hot spots identified here substantially increase the number of human hot spots available for analysis, absolute numbers remain small. Analyses of population data provide both a valuable resource for suggesting regions for future sperm-typing experiments and a fast, cheap and reliable method for identifying the large number of hot spots that may be necessary to unravel the complex, and perhaps highly heterogeneous, mechanisms underlying the recombination process.
Methods
Variation discovery.
As part of the National Heart, Lung, Blood Institute's Program for Genomic Applications, we resequenced 126 candidate genes involved in lipid metabolism, inflammation and blood pressure regulation to look for variation in 47 individuals from two populations: 23 unrelated European Americans from CEPH pedigrees (NA06990, NA07019, NA07348, NA07349, NA10830, NA10831, NA10842, NA10843, NA10842–NA10845, NA10848, NA10850–NA10854, NA10857, NA10858, NA10860, NA10861, NA12547, NA12548, NA12560) and 24 African Americans from the African-American Human Variation Panel (NA17101–17116 and NA17133–NA17140). All DNA samples are anonymous and are available to the public through Coriell Cell Repository. This study received a Human Subjects exemption from the University of Washington (no. 00-3300-X).
For a typical gene, we resequenced the genomic region spanning the longest reference transcript in LocusLink, including the exons and introns, as well as 2.5 kb upstream and 1.5 kb downstream on average. We designed overlapping primers for PCR to span the gene and then sequenced PCR products using Applied Biosystem's Big Dye sequencing protocol on ABI3700 and ABI3730 machines. Data analysts assembled the sequence data for each gene onto a reference genomic sequence using Phred and Phrap and edited the resulting alignments for accuracy using Consed. We then identified polymorphisms using PolyPhred 4.0 from pair-wise comparisons of individual sequence chromatograms in regions with an average Phred quality score >40. Analysts reviewed all polymorphisms identified by PolyPhred for false positives associated with features of the surrounding sequence or biochemical artifacts. If the polymorphism identified was an insertion-deletion polymorphism, analysts manually genotyped each sample and designed primers from the other strand to sequence past the polymorphism. Comparison of genotype calls from this protocol with calls on the TaqMan platform showed error rates to be < 1%. We deposited all genetic variation identified, and corresponding allele frequencies and genotypes, into GenBank and dbSNP.
To reduce computational demands, we ignored sites with minor allele frequency (MAF) <0.05. To reduce the chances of imprecise estimates from uninformative data, we analyzed only those genes that had, in both populations, at least 20 biallelic sites with MAF ≥0.05. We also excluded the two genes >100 kb in overall length (ITGA2, 106 kb; ITGA8, 206 kb), as the assumption of a single hot spot seemed least likely to hold for these longer genes. Of the initial pool of 126 genes, 74 genes met these requirements. A list of the 74 genes and GenBank accession numbers can be found in Supplementary Table 1 online. The molecular function of all 74 genes based on level 1 classification using PANTHER is listed in Supplementary Table 2 online.
Estimation of background rates of recombination and identification of hot spots.
To assess the support in the polymorphism data for a recombination hot spot, we used the 'product of approximate conditionals' model15. Because the haplotypic phase of our data is unknown, we incorporated the estimation of recombination parameters into the PHASE v2.1 software for haplotype estimation27,28 (see Supplementary Methods online). To compute BFs, we used the simple hot-spot model15, which assumes that there is a single hot spot of constant intensity λ. Crossovers occur as a Poisson process of constant rate r (per kilobase, per meiosis) outside the hot spot and of constant rate λr inside the hot spot (so λ = 1 corresponds to a constant recombination rate). The model has four parameters: the background population recombination parameter ρ (which equals 4Ner, where Ne is the effective population size); the intensity of the hot spot, λ; and the hot-spot center and width. The prior distribution for ρ was uniform on log10(ρ) in the range ρ = 10−5–106 per kb, spanning several orders of magnitude on either side of the expected average value for humans (0.4 per kb, assuming Ne = 10,000 and r = 1 cM Mb−1). We assumed a priori that the center of the hot spot was equally likely to be anywhere along the length of the sequence and that the width of the hot spot was ∼200–4,000 bp (specifically, that the width had a normal distribution, with mean 0 bp and standard deviation 2,000 bp, truncated to lie above 200 bp). For each population sample and each gene, we assumed that there was no recombination rate variation (i.e., λ = 1), that there was a 'warm spot' (1 < λ < 10) or that there was a hot spot (10 < λ < 100), with longer genes being less likely to show no variation. Specifically, for a gene of length L we assumed that λ = 1 with probability q = exp(−L/50,000); otherwise log10(λ) is uniformly distributed on (0, 2). This corresponds to a prior assumption that hot spots and warm spots each occur as a Poisson process of average rate 1 per 100 kb. We then used Markov Chain Monte Carlo (see Supplementary Methods online for further details) methods to obtain a sample of size 1,000 from the joint posterior distribution of all the parameters and the unknown haplotypes, given the genotype data G. From this, we estimated p10 = Pr(λ > 10 | G), the posterior probability of a hot spot given the data. The BFs in Figure 1 are Pr(G | λ > 10) / Pr(G | λ < 10), which, by Bayes theorem, equals p10 (q + 0.5(1 – q))/(1 – p10)(0.5(1 – q)). To allow for potential problems with convergence of the Markov Chain Monte Carlo algorithm, we ran the algorithm three times for each analysis, using different seeds for the pseudorandom number generator, and used the mean of the three estimates of p10 in computing the BFs. (We used the mean so that the BF is large only if all three estimates of p10 are large.) Similarly, to estimate the location and intensity of the hot spot, we used the posterior mean in each run and then used the median of the values obtained from the three runs. The CIs in Figure 3 and Supplementary Table 3 online are based on the sample of size 3,000 obtained by pooling the results from the three independent runs. Although combining results from independent runs in this way has no theoretical justification, we view it as a pragmatic way of avoiding gross errors due to poor convergence of the sampler in a single run. Supplementary Figure 2 online gives some idea of variability across runs.
Computational times for each application of the method varied from 4 min (C2 in African Americans) to 5.5 h (F5 in African Americans) on a 2.4-GHz processor with 4 Gb of RAM.
MHC data.
We obtained the MHC polymorphism data from the Jeffreys lab website. We divided the data into five nonoverlapping contiguous regions, each containing one of the hot spots identified by sperm typing. We analyzed each region independently, as described above.
Simulations.
We simulated the data used to produce Figure 2b,c and Supplementary Figure 1 online using coalescent-based software29. For Figure 2b,c, we assumed that an ancestral population of size 9,000 split 2,800 generations ago into two populations, whose sizes instantaneously changed to 9,000 and 10,000, respectively. The first population experienced exponential growth from 9,000 to 10,000 individuals starting 2,000 generations ago. The second population experienced a bottleneck 400–600 generations ago, during which its size reduced to 500 individuals, before returning to its prebottleneck size. Finally, 20 generations ago, one-way gene flow started from the second population to the first, at a rate of 0.25 migrants per generation. These parameters were provided by J. Akey and were chosen to match approximately the real data in various summary statistics, including the divergence between the populations (measured by FST), and Tajima's D in each population (J. Akey, personal communication). For the data used to produce Supplementary Figure 1 online, we simulated data for two populations independently, under two different demographic scenarios: expansion, and bottleneck followed by expansion. For each scenario, we assumed an ancestral population size of 8,600, which grew exponentially from 500 generations ago to a present size of 100,000. For the bottleneck scenario, the population was also assumed to undergo a bottleneck 900–2,000 generations ago, during which its size was reduced to 860 individuals. After the bottleneck, the population returned to its prebottleneck size of 8,600 individuals before expanding as in the expansion scenario. The parameters of the bottleneck simulation were chosen to reduce expected SNP density to ∼60% of the levels of the expansion scenario, approximately mimicking the reduction in our European American (as compared with African American) samples.
In each case, we simulated data for 74 genes that matched the data on the 74 real genes with respect to numbers of individuals, numbers of SNPs with MAF >5%, physical length and background levels of LD. This last was accomplished by using kρA as the value of ρ = 4N0r for the simulations, where ρA is the estimated value of ρ from the real data from African American samples and k is a constant chosen to account for the fact that the software requires ρ to be entered in terms of the current population size N0 rather than the effective population size (we used k = 4/3 for Fig. 2; k = 10 for Supplementary Figure 1 online). For the simulations in Figure 2, we also matched the number of SNPs that were shared across populations, by first simulating a large number of total SNPs and then randomly selecting a subset of these SNPs such that the number of SNPs that exceeded MAF >0.05 in (i) both populations, (ii) the first population only or (iii) the second population only matched our real data. For each simulated gene, gene conversion was assumed to occur uniformly across the gene, with average tract length of 250 bp and at a rate of zero, one or ten times the crossover rate, with roughly equal numbers of genes in each rate category30.
To create data with recombination hot spots (Fig. 2c and Supplementary Fig. 1 online) we used a previously described algorithm15. For each gene, we chose the location and intensity of the hot spot to match the corresponding estimates from the real data from European American (for Fig. 2c) or African American samples (for Supplementary Fig. 1 online).
LR computations.
For genes showing substantive evidence for a hot spot in only one population sample, we computed the LR to test the null hypothesis that the intensity of the hot spot is the same in each population. We first fixed the hot-spot location at the position estimated from the sample with substantive evidence for a hot spot and used a Markov Chain Monte Carlo scheme as above to sample from the posterior distribution for log10(λ), independently for each sample, using a uniform prior distribution on log10(λ) = 0–2 and the prior distribution on ρ given above. We then used kernel density estimation (from the R statistical package) to estimate the posterior density for log10(λ) for each sample. Because of our uniform prior distribution, this posterior density is proportional to the likelihood. We computed LRs under the assumption that the data in the two samples were independent and were therefore equal to LRA(λA) LRE(λE) / LRA(λ0) LRE(λ0), where LRA(.) is the likelihood for the African American sample, LRE(.) is the likelihood for the European American sample, λA is the value of λ that maximizes LRA(λ), λE is the value of λ that maximizes LRE(λ) and λ0 is the value of λ that maximizes LRA(λ)LRE(λ). All maximizations were done over a dense grid of λ values. In some cases, the likelihood seemed to be maximized above log10(λ) = 2, in which case we repeated the process with a uniform prior distribution on log10(λ) = 0–3. We calculated P values with the assumption that under the null hypothesis, 2log(LR) has an asymptotic χ2 distribution on 1 degree of freedom.
Sequence motif search.
We searched each gene for the sequence motifs hypothesized to affect recombination rates (listed in Supplementary Table 4 online) and for microsatellites and 'words' (with alphabet A,C,G,T) to determine whether they are over-represented in the hot-spot regions. Microsatellites were defined by a minimum number of repeats for each unit size: ten repeats for a mononucleotide repeat, six repeats for a dinucleotide repeat, five repeats for a trinucleotide repeat, three repeats for a tetranucleotide repeat or greater. We searched the sequence of each gene for words as long as 15 letters. To determine whether sequence motifs, microsatellites or words were significantly associated with hot spots, we counted the number of occurrences in the hot-spot region and the number of occurrences outside the hot-spot region for each population separately. We then carried out a boot-strap analysis to calculate the P values for tests of significance. For the boot-strap procedure, for each gene, we independently simulated the presence or absence of a hot spot with the probability of a hot spot being the number of genes with BF > 10 for each population divided by 74. For each gene assigned a hot spot in the simulation, we chose the hot-spot width at random from the empirical distribution of width estimates in each population. The position of the hot spot in the gene was selected uniformly along the DNA sequence, conditional on the hot spot being contained entirely within the DNA sequence. We counted the number of occurrences of the sequence motifs, microsatellites or words in the simulated hot spots. The simulations were repeated 10,000 times. We carried out a similar boot-strap analysis to determine whether G+C content was significantly associated with the presence of hot spots.
To investigate association between the estimated background recombination rate (ρ) and G+C content, nucleotide diversity, repetitive elements, sequence motifs and map-based estimates of recombination rate, we carried out multiple linear regressions of log(ρ) against these variables for each population sample separately, using R. We calculated nucleotide diversity as the average number of pairwise sequence differences across each gene (Supplementary Table 1 online). Repetitive elements were identified using RepeatMasker Map-based estimated of recombination rates were deCode estimates18 obtained from the University of California Santa Cruz Genome Browser.
URLs.
The Program for Genomic Applications from SeattleSNPs is available at http://pga.gs.washington.edu. Phred, Phrap and Consed are available at http://www.phrap.org. Polyphred 4.0 is available at http://droog.mbt.washington.edu./PolyPhred.html. PANTHER is available at http://panther.appliedbiosystems.com/about.jsp. PHASEv2.1 is available at http://www.stat.washington.edu/stephens/software.html. The Jeffreys lab website is http://www.le.ac.uk/ge/ajj/HLA/index.html. The R statistical package is available at http://www.R-project.org. The University of California Santa Cruz Genome Browser is available at http://genome.ucsc.edu/cgi-bin/hgGateway. RepeatMasker is available at http://www.repeatmasker.org/.
Note: Supplementary information is available on the Nature Genetics website.
References
de Massy, B. Distribution of meiotic recombination sites. Trends Genet. 19, 514–522 (2003).
Pritchard, J.K. & Przeworski, M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69, 1–14 (2001).
Clark, A.G. et al. Linkage disequilibrium and inference of ancestral recombination in 538 single-nucleotide polymorphism clusters across the human genome. Am. J. Hum. Genet. 73, 285–300 (2003).
Jeffreys, A.J., Ritchie, A. & Neumann, R. High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum. Mol. Genet. 9, 725–733 (2000).
Jeffreys, A.J., Kauppi, L. & Neumann, R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29, 217–222 (2001).
Schneider, J.A., Peto, T.E., Boone, R.A., Boyce, A.J. & Clegg, J.B. Direct measurement of the male recombination fraction in the human beta-globin hot spot. Hum. Mol. Genet. 11, 207–215 (2002).
Arnheim, N., Calabrese, P. & Nordborg, M. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am. J. Hum. Genet. 73, 5–16 (2003).
Daly, M.J., Rioux, J.D., Schaffner, S.F., Hudson, T.J. & Lander, E.S. High-resolution haplotype structure in the human genome. Nat. Genet. 29, 229–232 (2001).
Patil, N. et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294, 1719–1723 (2001).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Wang, N., Akey, J.M., Zhang, K., Chakraborty, R. & Jin, L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 71, 1227–1234 (2002).
Phillips, M.S. et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat. Genet. 33, 382–387 (2003).
Wall, J.D. & Pritchard, J.K. Assessing the performance of the haplotype block model of linkage disequilibrium. Am. J. Hum. Genet. 73, 502–515 (2003).
Chakravarti, A. et al. Nonuniform recombination within the human beta-globin gene cluster. Am. J. Hum. Genet. 36, 1239–1258 (1984).
Li, N. & Stephens, M. A new multilocus model for linkage disequilibrium, with application to exploring variations in recombination rate. Genetics 165, 2213–2233 (2003).
Li, N. Modeling and inference for linkage disequilibrium and recombination. (PhD Thesis, Department of Biostatistics, University of Washington, 2003)
Gerton, J.L. et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97, 11383–11390 (2000).
Kong, A. et al. A high-resolution recombination map of the human genome. Nat. Genet. 31, 241–247 (2002).
Cullen, M., Perfetto, S.P., Klitz, W., Nelson, G.W. & Carrington, M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am. J. Hum. Genet. 71, 759–776 (2002).
Wall, J.D., Frisse, L.A., Hudson, R.R. & Di Rienzo, A. Comparative linkage-disequilibrium analysis of the beta-globin hotspot in primates. Am. J. Hum. Genet. 73, 1330–1340 (2003).
Ptak, S.E. et al. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. (in the press).
Nachman, M.W., Bauer, V.L., Crowell, S.L. & Aquadro, C.F. DNA variability and recombination rates at X-linked loci in humans. Genetics 150, 1133–1141 (1998).
Ptak, S.E., Voelpel, K. & Przeworski, M. Contrasting large scale and local recombination rates in humans. Genetics (in the press).
Fullerton, S.M., Carvalho, A.B. & Clark, A.G. Local rates of recombination are positively correlated with GC content in the human genome. Mol. Biol. Evol. 18, 1139–1142 (2001).
Petes, T.D. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369 (2001).
Fullerton, S.M. et al. Polymorphism and divergence in the beta-globin replication origin initiation region. Mol. Biol. Evol. 17, 179–188 (2000).
Stephens, M., Smith, N.J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Stephens, M. & Donnelly, P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 (2003).
Hudson, R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002).
Frisse, L. et al. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69, 83–843 (2001).
Acknowledgements
We thank members of the SeattleSNPs team and the laboratory of D.A.N. (M. Ahearn, B. Borrayo, E. Calhoun, M. Chung, S. Da Ponte, L. Daniels, M. Daniels, C. Hastings, B. Howie, P. Keyes, P. Lee, B. Livingston, R. Mackelprang, M. Montoya, C. Nguyen, D. Nguyen, C. Poel, N. Rajkumar, P. Robertson, W. Schackwitz, T. Shaffer, A. Sherwood, K. Sherwood, J. Sloan, R. Torskey, E. Toth, L. Witrack. M. Wong and Q. Yi) for their efforts in variation discovery and M. Przeworski for critical reading of this manuscript. This work was supported by grants from the National Heart Lung and Blood Institute Program for Genomic Applications (D.A.N. and M.J.R.), the National Institute for Environmental Health (D.A.N and M.J.R.) and the National Human Genome Research Institute (M.S.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Bayes Factors (BFs) summarizing the strength of the evidence for a hotspot in each gene, and each population sample, for simulated data with and without recombination hotspots. (PDF 127 kb)
Supplementary Fig. 2
Estimated intensity and location of hotspots, based on African-American sample (red) and European sample (green), for all genes. (PDF 127 kb)
Supplementary Table 1
Gene name, location, size, and number of SNPs (≥5% minor allele frequency), nucleotide diversity (π), and G+C content. (PDF 6 kb)
Supplementary Table 2
Molecular function based on PANTHER terms. (PDF 4 kb)
Supplementary Table 3
Estimated background recombination rate (ρ) per kilobase; map-based estimates (cM/Mb); and estimated average recombination rate per kilobase for each gene. (PDF 9 kb)
Supplementary Table 4
Sequence motifs and their occurrences within and outside hotspots. (PDF 6 kb)
Supplementary Methods
Further details on Markov Chain Monte Carlo procedure and caveats on interpretation of LRs. (PDF 9 kb)
Rights and permissions
About this article
Cite this article
Crawford, D., Bhangale, T., Li, N. et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet 36, 700–706 (2004). https://doi.org/10.1038/ng1376
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1376
This article is cited by
-
Genome-wide characterization of non-reference transposable element insertion polymorphisms reveals genetic diversity in tropical and temperate maize
BMC Genomics (2017)
-
ARG-walker: inference of individual specific strengths of meiotic recombination hotspots by population genomics analysis
BMC Genomics (2015)
-
High-resolution linkage map for two honeybee chromosomes: the hotspot quest
Molecular Genetics and Genomics (2014)
-
Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer
Nature Genetics (2014)
-
Meta-analysis identifies four new loci associated with testicular germ cell tumor
Nature Genetics (2013)