Abstract
Telomere length is a crucial factor in senescence1,2,3, but it has not been determined whether animals with long telomeres live longer than those with normal-length telomeres in the isogenic background of a given species. Here we show the effect of long telomeres on lifespan in the nematode Caenorhabditis elegans. We examined the effect of telomere length on lifespan by overexpressing HRP-1, a telomere-binding protein, which gradually increased telomere length in worms. Worms with longer telomeres lived longer. We confirmed that the extension of lifespan was due to the increased telomere length, and not to the overexpression of HRP-1 per se, by examining the lifespans of nontransgenic progeny of the transgenic worms, who retained the longer telomeres. The lifespan-extending effect of long telomeres was dependent on daf-16. The number of germ stem cells was not affected in worms with long telomeres, indicating that the telomere effect on lifespan is independent of germ stem cell cycling. Worms with long telomeres were more resistant to heat stress. Taken together, our results suggest that signaling may be initiated in postmitotic somatic cells by telomere length to regulate organismal lifespan.
Similar content being viewed by others
Main
Chromosomes in linear forms are doomed to be shortened by the nature of DNA replication and, unless there is a specific protection mechanism, limit the number of cell divisions1,2. The telomere hypothesis explains how telomere length limits cellular senescence3. Knockout mice lacking telomerase activity experience premature aging4. But the mechanisms acting at the level of replicative aging may not be applicable to the aging process and organismal lifespan, because most somatic cells are postmitotic. Cells that overexpress TERT live longer and have slower cellular senescence5, but this phenomenon may simply result from delaying the crisis point provoked by critically short telomeres. Telomere length was suggested not to be a primary factor in determining organismal lifespan6, but because the animals compared were not in an identical genetic background, other factors may have affected their lifespan. Therefore, the question of whether animals with long telomeres live longer is still open.
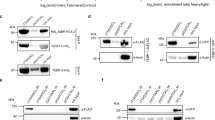
To address this question, we used an extensively inbred strain (N2) of the nematode C. elegans in which any individual worm is considered to have an isogenic genetic background. C. elegans has typical telomere repeat sequences with an average length of 4–9 kb (ref. 7). As no telomere-binding protein that regulates telomere length has been reported in C. elegans, we first identified proteins that specifically bound single-stranded telomeric DNA by affinity purification and MALDI-TOF analysis. We identified HRP-1, a protein encoded by F42A6.7a, which shares high sequence homology with human RNP A1 and yeast GBP2 (Fig. 1a). These two proteins bind single-stranded telomeric DNA and are involved in telomere length regulation8,9,10. We confirmed that HRP-1 specifically bound to G-rich single-stranded telomeric DNA (Fig. 1b). HRP-1 binding was efficiently inhibited by the (TTAGGC)4 cold competitor but not by a nonspecific single-stranded competitor. The C-rich sequence (GCCTAA)4 and the double-stranded telomeric DNA (TTAGGC)4D were not efficiently bound by HRP-1. In a gel-shift assay using (TTAGGC)2–4 telomeric repeats as probes, we found that 2.5 (TTAGGC) repeats was the minimum for HRP-1 binding. The affinity of HRP-1 binding was markedly higher when we used four (TTAGGC) repeats as a probe, consistent with previous reports that RNP A1 binding to telomeric DNA is length-dependent9 (Fig. 1c). We found that HRP-1 was expressed in all nuclei in somatic cells in a green fluorescent protein (GFP) reporter assay (Fig. 1d). Using antibodies against HRP-1, we found that HRP-1 was localized at the ends of the chromosomes of the germ cells (Fig. 1e). These results show that HRP-1 is expressed in most germ and somatic cells and is localized mainly at the telomeres in vivo.
(a) Amino acid sequence alignment of HRP-1 homologs. Human RNP A1 and yeast GBP2 are putative homologs. Identical amino acids are shaded. (b) Specificity of HRP-1 binding. Gel-shift assays were carried out using purified HRP-1 with the 32P-end-labeled (TTAGGC)4 probe in the presence of different amounts of unlabeled (TTAGGC)4 (lanes 2–6) and nonspecific oligonucleotides (lanes 7–11). The relative amounts of the competitors added are indicated. The bottom panel shows free probes. (c) Length-dependent and G-rich single-stranded telomere binding of HRP-1. Gel-shift assays were carried out with 32P-end-labeled probes. Lane 1, (TTAGGC)2; lane 2, (TTAGGC)2.5; lane 3, (TTAGGC)3; lane 4, (TTAGGC)4, lane 5, (GCCTAA)4; lane 6, (TTAGGC)4D. 4D denotes a double-stranded form. (d) Ubiquitous expression of hrp-1 in the somatic cells, as shown by GFP reporter fluorescence. (e) Localization of HRP-1 at the chromosome ends, as shown by antibody staining of HRP-1 in a germ cell. Blue staining is DAPI, and green is the HRP-1 signal. There are three punctuated spots of HRP-1 at the chromosome ends.
To examine whether HRP-1 is involved in telomere regulation in vivo, we established a transgenic line that overexpressed an hrp-1:gfp transgene (Fig. 2a,b). Southern-blot analysis showed that telomere length increased in successive generations of this transgenic line (Fig. 2c). We observed the same phenomenon in the mut-7 mutant background (Fig. 2c). The mut-7 (pk204) mutation prevents germline suppression of transgenes, making possible the expression of the transgene in the germ line and early embryos11. We also examined whether loss of overexpression of HRP-1 caused a decrease in telomere length in successive generations, taking advantage of the fact that the transgenic line that we used was not an integrated line. We obtained progeny that had lost the transgene in the F25 generation and propagated the nontransgenic worms over several generations (Fig. 2a,b). Telomere length in the nontransgenic progeny of F25 worms gradually decreased in successive generations (Fig. 2c). We concluded from these results that HRP-1 is involved in telomere length regulation in vivo.
(a) Experimental procedures. +, worms that retain the extrachromosomal array; −, worms that have lost the extrachromosomal array. For example, F25–2 worms are the grandchildren of F25 transgenic worms that lost the transgene two generations ago. (b) Western-blot analysis using polyclonal antibody to HRP-1. The upper band is HRP-1–GFP, and the two lower bands are two alternative forms of endogenous HRP-1. Only F25 worms retain HRP-1–GFP; their progeny have lost the transgene. Prolonged exposure of the same blot did not show any sign of residual HRP-1–GFP in F25–2 worms (data not shown). (c) Overexpression of hrp-1 increases telomere lengths. The two left panels are in the N2 background, and the two middle panels are in the mut-7 background. In successive generations, the telomeres became longer in both genetic backgrounds. In the wild-type N2 background, telomeres of the F25 progeny were 2 kb longer than those of control worms. In the mut-7 background, telomeres of the F20 progeny were 1.5 kb longer than those of control worms. After the transgenic worms lost the transgene, telomeres shortened in successive generations, as shown by comparing the F25–2 and F25–7 worms. The right panel shows telomere lengths in the daf-16 background. Telomeres became longer in this genetic background also. The locations of the DNA size markers are indicated to the left of each panel.
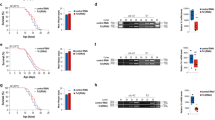
We next tested whether long telomeres increased the lifespan of worms containing somatic cells at their postmitotic stage. To exclude the possibility that overexpression of HRP-1 per se affected the lifespan of the worms, we examined worms that had lost the transgene but still had long telomeres (F25–1 and F25–2 progeny, which came from F25 worms that had lost the transgene; Fig. 2c and data not shown). These worms had no apparent abnormalities in development or behavior (data not shown). F25–2 progeny (n = 327) had a mean lifespan that was 19.3% longer than that of N2 worms (n = 361; P < 0.0001; Fig. 3a). Telomeres in F25–7 progeny were longer than those of control worms but shorter than those of F25–2 worms (Fig. 2c); telomere length increased by 10.0% in F25–7 worms (n = 56, P = 0.0018; Fig. 3a). F25–18 worms, whose telomeres were not longer significantly longer than those of N2 controls, did not have a significantly longer mean lifespan (3.1% increase, n = 62, P = 0.82; Fig. 3a). The maximum lifespan was also longer in F25–2 worms. Lipofuscin accumulation, a typical aging marker, began 2–3 d later in F25–2 worms than in N2 controls (Fig. 3c), indicating that telomere length has an effect not only on the mean lifespan but also on the rate of aging. We obtained similar results from experiments on the mut-7 mutant background (Fig. 3b and data not shown). mut-7 mutant worms tended to live longer than wild-type worms. We concluded from these data that the long telomeres, and not the overexpression of HRP-1, increased the lifespan of the worms.
(a) Lifespan in the N2 background. The table shows the numbers of worms examined, their mean lifespan (in d) and P values for each generation compared with the N2 control. F25–2 and F25–7 worms, but not F25–18 worms, had significantly longer mean lifespan than N2 control worms. (b) Lifespan in the mut-7 background. F25–2 and F25–7 worms had significantly longer mean lifespans, but F25–18 worms had only a marginal increase in the mean lifespan. The table shows the numbers of worms examined, their mean lifespan (in d) and P values for each generation compared with the N2 control. (c) Lipofuscin accumulation is delayed in F25–2 worms. The arrowheads indicate lipofuscin accumulation in the gut. The number of days passed after reaching adulthood is indicated above each photo. Control worms began to accumulate lipofuscin autofluorescence at day 2, whereas the F25–2 worms began to show accumulation at day 4. After day 4, there was no significant difference in the intensity of the lipofuscin autofluorescence between F25–2 and control worms.
Animals that live longer are more resistant to oxidative stress and heat shock12,13,14,15. We examined the resistance of worms with long telomere to various stresses. F25–2 worms, which have long telomeres, but not F3–2 or F25–25 worms, which do not have long telomeres, had significantly greater survival rates at 35 °C heat shock (Fig. 4a,b). But F25–2 worms were not significantly more resistant to paraquat, a representative oxidative stress (data not shown), implying that there is a correlation between telomere length and resistance to a subset of environmental stresses.
(a) Worms with long telomeres are more resistant to heat shock. Five-day-old N2 and F25–2 worms were transferred to 35 °C and survival was scored at various time points after treatment (n = 120 for each strain at each time point). The same experiments were repeated at least three times, and cumulative results of all experiments are shown. (b) Comparison of resistance to heat shock of 35 °C for 8 h in worms with telomeres of various lengths. F3–2 and F25–25 worms were examined as examples of worms with normal-length telomeres. Whereas F3–3 (n = 189) and F25–25 (n = 200) worms were not significantly more resistant to heat shock (P = 0.176 and 0.204, respectively), F25–2 worms (n = 180) were (P < 0.05). The number of N2 control worms examined was 175. (c) The effect of long telomeres on lifespan is dependent on daf-16. The mean lifespan of daf-16 (F25–2) worms was not significantly different from that of the control daf-16 mutant strain. In this experiment, N2 and N2 (F25–2) worms were also examined as controls.
daf-16 is a key component in a main pathway that determines the lifespan of the nematode16,17. Nuclear translocation of DAF-16 increases transcription of stress-response genes18. To determine whether the lifespan extension attributable to long telomeres is relevant to the daf-16 signaling pathway, we examined the lifespan of daf-16 worms with long telomeres (F25–2 progeny) and found that it was not longer than that of control daf-16 mutant worms (Fig. 4c). This result indicates that the effect of telomere length on lifespan is dependent on the functional daf-16 gene. To examine the possibility that the telomere signal is transmitted through the germ cells, in which signaling for lifespan extension is dependent on daf-16 (ref. 19), we determined whether germ stem cells in worms with long telomeres proliferate more slowly. We observed no significant difference in the number of germ stem cells, indicating that the effect of long telomeres on lifespan is independent of cell cycling of the germ stem cells. Our results indicate that the effect of long telomeres on organismal lifespan may be, at least for the nematode, a somatic event.
Of the C. elegans genes identified so far whose mutations make worms live longer, only one gene, clk-2, was reported to affect telomere length, with some controversy20,21,22. It would be difficult to determine whether long telomeres extend lifespan in these worms, because the clk-2 mutation causes many other pleiotropic effects, such as a defective DNA checkpoint function and slow metabolism23. In contrast, our results clearly establish a causal relationship between long telomeres and extended lifespan. Why do worms with long telomeres live longer? It is formally possible that the long telomere is a cellular damage signal that induces stress resistance in the soma. Another possibility is that the long telomeres exert a position effect on genes at the subtelomeric regions. This possibility could be investigated further by microarray experiments. A third possibility is that proteins binding to the telomeres have a quantitative effect on a signaling pathway in determining the metabolism of the cell and the organism. Determining the effect on lifespan of mutations in the telomere-binding proteins that do not affect the telomere length could help to explore this possibility.
In summary, we report a new role for telomere length in organismal lifespan. We propose that telomere length is not only essential for regulating the onset of senescence at the cellular level, but also important in determining the lifespan of a whole animal. Nematodes with long telomeres developed at a normal rate (data not shown) and lived longer at their adult stage; it would be interesting to determine whether the same is true for mammals. Notably, it was recently reported that people with long telomeres, although with independent genetic backgrounds, have lower mortality than those with short telomeres24. Lengthening the telomeres in adult somatic cells by a means that does not cause side effects could help mammals, including humans, with relatively short telomeres to live longer.
Methods
Affinity chromatography and analysis of purified proteins.
We prepared nematode embryonic nuclear extract as previously described25. We used agarose beads with covalently linked streptavidin- and biotin-labeled C. elegans single-strand telomeric DNA repeats to purify proteins that specifically bound the single-strand telomeric DNA. We carried out mass spectrometry followed by one- or two-dimensional gel electrophoresis to identify the purified proteins. We identified many candidate proteins, including HRP-1 in both one- and two-dimensional analysis, indicating that this protein may be a main single-stranded telomere binding protein.
HRP-1 expression studies.
We subcloned the cDNA of the open reading frame of F42A6.7a into the pRSET-A vector, introduced this expression construct into the BL21 strain and purified overexpressed protein using Ni-NTA agarose beads (Qiagen). Antibodies specific to the bacterially overexpressed HRP-1 were produced (by Takara Korea) in a rabbit and the antigen was affinity-purified. We carried out western-blot analysis according to standard procedures. We visualized signals using an enhanced chemiluminescense kit (Amersham). We collected gonads from adult worms for immnuohistochemical analysis. After treating them with appropriate primary and secondary antibodies, we observed the fluorescence of the gonads using a fluorescence microscope (Carl Zeiss). We used DAPI to visualize the chromosomes. To observe the expression of a GFP reporter, we amplified genomic DNA containing the gene F42A6.7a by PCR and cloned into the pPD95.77 vector (gift from A. Fire, Stanford University). We microinjected the GFP reporter construct as described26. We observed fluorescence of the transgenic worms using a fluorescence microscope equipped with Nomarski optics (Carl Zeiss).
Electrophoretic mobility shift assays.
We obtained DNA probes and competitors used in these assays from BIONEER, ZENOTECH and BIONIX. The probes were radioactively labeled using γ-[32P]-ATP (Amersham) and purified on 15% polyacrylamide gels. For gel shift assays, we purified HRP-1 protein from bacteria and incubated it in 20 μl of the binding buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM dithiothreitol, 50 mM NaCl and 5% glycerol) and 500 ng of poly(dI-dC) on ice for 10 min. We added 0.25 ng of the end-labeled DNA probe to the premixture, incubated it at room temperature for 15 min and then separated it on 8% nondenaturing polyacrylamide gels. We detected signals using X-ray films or Fuji imaging plates. For competition assays, we added the cold competitors to the purified protein before adding the labeled probe.
Lipofuscin autofluorescence measurement.
We measured gut lipofuscin autofluorescence of F25–2 (n = 149) and N2 control worms (n = 146) as described27. We photographed gut autofluorescence using a 525-nm band-pass filter. Images had the same exposure time, and we collected them without any automatic gain control to preserve the relative intensity of different samples.
Southern-blot analysis of telomere length.
We carried out Southern-blot analysis of telomeres in the N2 and mut-7 backgrounds. We purified C. elegans genomic DNA, from various strains including control worms and worms overexpressing HRP-1, using the puregene DNA purification kit (GENTRA). We digested 5 μg of genomic DNA with HinfI, separated it on a 0.7% agarose gel and subjected it to Southern hybridization with a radioactively labeled DNA probe made of (TTAGGC)40. We examined the results using a phosphorimaging device (Fuji). Because multicopy transgenes tend to be silenced in the germ cells, we made sure that the transgene was expressed in the embryonic germ cells by observing GFP fluorescence in every cell at early embryonic stages (data not shown). We hypothesized that although the transgene product might not act in the germ cells, its expression in the germline precursor cells at the embryonic stage would be enough to function, at least partially, as a telomere-length regulator.
Lifespan and stress assays.
We measured the lifespan of the nematodes at 20 °C. We transferred adult worms to a new plate and removed them after allowing them to lay eggs for 12 h. We then examined the lifespan of the progeny. The day 0 was defined as the day when the adults were removed. After 4 d, we transferred five L4-stage worms to new plates every day until they did not lay any more eggs. We then transferred them to new plates every other day. We regarded the worm as dead if it did not respond to a gentle touch. Worms that died of causes other than aging, such as sticking to the plate walls, bursting in the vulva region or forming worm bags, were excluded from our analysis. We used the log-rank (Mantel-Cox) test to evaluate the significance of the differences in mean lifespan. We calculated P values comparing mean lifespans with either wild-type control in all experiments or wild-type control in individual experiments, as indicated in figure legends. For the heat-shock assay, we transferred 5-d-old worms to new plates and shifted them to 35 °C. We counted live worms at specific time points. For the oxidative stress assay, we added 60 mM paraquat to the plates and counted live worms at specific time points. Each experiment was repeated at least twice.
Examination of germ stem cells.
To examine the numbers of germ stem cells in the gonads of the worms, we isolated the gonads and visualized the nuclei of the germ cells by DAPI staining. We counted the nuclei of the germ stem cells according to the criteria of shape and appearance in DAPI staining. The average numbers of germ stem cells in each gonad arm of young adult worms 1–3 d after reaching adulthood were 169.1 and 167.4 for the F25–2 (n = 56) and the N2 (n = 56) control worms, respectively (P = 0.79).
References
Krupp, G., Klapper, W. & Parwaresch, R. Cell proliferation, carcinogenesis and diverse mechanisms of telomerase regulation. Cell Mol. Life Sci. 57, 464–486 (2000).
Morin, G.B. Telomere control of replicative lifespan. Exp. Gerontol. 32, 375–382 (1997).
Harley, C.B., Vaziri, H., Counter, C.M. & Allsopp, R.C. The telomere hypothesis of cellular aging. Exp. Gerontol. 27, 375–382 (1992).
Rudolph, K.L. et al. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712 (1999).
Bodnar, A.G. et al. Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352 (1998).
Hemann, M.T. & Greider, C.W. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 28, 4474–447 (2000).
Wicky, C. et al. Telomeric repeats (TTAGGC)n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 93, 8983–8988 (1996).
Lin, J.J. & Zakian, V.A. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 22, 4906–4913 (1994).
LaBranche, H. et al. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19, 199–202 (1998).
Fiset, S. & Chabot, B. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 29, 2268–2275 (2001).
Ketting, R.F., Haverkamp, T.H., van Luenen, H.G. & Plasterk, R.H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141 (1999).
Lithgow, G.J., White, T.M., Hinerfeld, D.A. & Johnson, T.E. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J. Gerontol. 49, B270–276 (1994).
Lithgow, G.J., White, T.M., Melov, S. & Johnson, T.E. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. USA 92, 7540–7544 (1995).
Honda, Y. & Honda, S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 13, 1385–1393 (1999).
Larsen, P.L. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 90, 8905–8909 (1993).
Lin, K., Dorman, J.B., Rodan, A. & Kenyon, C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322 (1997).
Ogg, S. et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999 (1997).
Lin, K., Hsin, H., Libina, N. & Kenyon, C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28, 139–145 (2001).
Hsin, H. & Kenyon, C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366 (1999).
Benard, C. et al. The C. elegans maternal-effect gene clk-2 is essential for embryonic development, encodes a protein homologous to yeast Tel2p and affects telomere length. Development 128, 4045–4055 (2001).
Lim, C.S., Mian, I.S., Dernburg, A.F. & Campisi, J. C. elegans clk-2, a gene that limits life span, encodes a telomere length regulator similar to yeast telomere binding protein Tel2p. Curr. Biol. 11, 1706–1710 (2001).
Ahmed, S., Alpi, A., Hengartner, M.O. & Gartner, A. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 11, 1934–194. (2001).
Benard, C. & Hekimi, S. Long-lived mutants, the rate of aging, telomeres and the germline in Caenorhabditis elegans. Mech. Ageing Dev. 123, 869–880 (2002).
Cawthon, R.M., Smith, K.R., O'Brien, E., Sivatchenko, A. & Kerber, R.A. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361, 393–395 (2003).
Kwon, J.Y. et al. Caenorhabditis elegans mediator complexes are required for developmental-specific transcriptional activation. Proc. Natl. Acad. Sci. USA 96, 14990–14995 (1999).
Mello, C.C., Kramer, J.M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991).
Garigan, D. et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161, 1101–1112 (2002).
Acknowledgements
We thank C. Kenyon for her comments and invaluable advice on this project, A. Fire for the vectors and CGC for the strains. This project was supported by the Molecular Aging Research Center Grant (HPEB, Korea).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Joeng, K., Song, E., Lee, KJ. et al. Long lifespan in worms with long telomeric DNA. Nat Genet 36, 607–611 (2004). https://doi.org/10.1038/ng1356
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1356
This article is cited by
-
Telomere Length is a Susceptibility Marker for Tasmanian Devil Facial Tumor Disease
EcoHealth (2020)
-
Splicing factors control C. elegans behavioural learning in a single neuron by producing DAF-2c receptor
Nature Communications (2016)
-
Rapid growth accelerates telomere attrition in a transgenic fish
BMC Evolutionary Biology (2015)
-
Heritability of telomere length in the Zebra Finch
Journal of Ornithology (2015)
-
Telomere dynamics in a long-lived bird, the barnacle goose
BMC Evolutionary Biology (2012)