Abstract
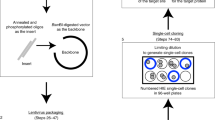
RNA interference (RNAi) has recently emerged as a specific and efficient method to silence gene expression in mammalian cells either by transfection of short interfering RNAs (siRNAs; ref. 1) or, more recently, by transcription of short hairpin RNAs (shRNAs) from expression vectors and retroviruses2,3,4,5,6,7,8,9,10. But the resistance of important cell types to transduction by these approaches, both in vitro and in vivo11, has limited the use of RNAi. Here we describe a lentiviral system for delivery of shRNAs into cycling and non-cycling mammalian cells, stem cells, zygotes and their differentiated progeny. We show that lentivirus-delivered shRNAs are capable of specific, highly stable and functional silencing of gene expression in a variety of cell types and also in transgenic mice. Our lentiviral vectors should permit rapid and efficient analysis of gene function in primary human and animal cells and tissues and generation of animals that show reduced expression of specific genes. They may also provide new approaches for gene therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
14 May 2007
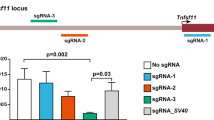
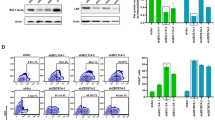
The authors wish to remove panels b, d, and e of Figure 2 because they were falsified or fabricated. Figure 2 shows the functional silencing of genes in primary cells by lentiviral-mediated RNAi. The authors showed that primary activated T cells can be successfully transduced by lentivirus and that RNAi-targeted surface markers are downregulated in transduced cells (Fig. 2a). This knockdown leads to a deficiency in T cell proliferation in response to IL-2 (Fig. 2c). These conclusions remain intact. Figure 2b demonstrates the transduction of bone marrow–derived dendritic cells and the knockdown of bim expression. Figure 2d shows the transduction of hematopoietic stem cells (HSCs) and the expression of GFP marker in splenocytes from chimeras derived from transduced HSCs. Figure 2e shows the reduction in CD8+ cells harvested in CD8-knockdown bone marrow chimeras. However, the data supporting the transduction of dendritic cells or HSCs with the lentiviral vector pLL3.7 and the resulting knockdown has been determined to be falsified or fabricated, invalidating the conclusions drawn from these panels (Fig. 2b,d,e). We have no reason to believe that the other results and interpretations in this paper need to be corrected. This correction follows an investigation by the Massachusetts Institute of Technology into scientific misconduct by Luk Van Parijs, the communicating author of the paper. The investigation also found that Dr. Van Parijs was solely responsible for the scientific misconduct that resulted in the falsified or fabricated data in this paper. In addition, the authors wish to revise a Corrigendum regarding this paper that was published at Dr. Van Parijs' request (Nat. Genet. 34, 231; 2003). The Corrigendum contained a revised author list that mistakenly omitted Mingdi Zhang, an author on the original paper. The complete author list is as follows: Douglas A. Rubinson, Christopher P. Dillon, Adam V. Kwiatkowski, Claudia Sievers, Lili Yang, Johnny Kopinja, Dina L. Rooney, Mingdi Zhang, Melanie M. Ihrig, Michael T. McManus, Frank B. Gertler, Martin L. Scott & Luk Van Parijs.That Corrigendum also acknowledged partial support from the Broad Institute for the work reported in the paper. The Broad Institute in fact did not support any of that work, and the acknowledgment of its support was mistaken.Editor’s note: all of the authors, with the exception of Luk Van Parijs, agree with the wording of this Corrigendum.
References
Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Lee, N.S. et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20, 500–505 (2002).
Brummelkamp, T.R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
McManus, M.T., Petersen, C.P., Haines, B.B., Chen, J. & Sharp, P.A. Gene silencing using micro-RNA designed hairpins. RNA 8, 842–850 (2002).
Miyagishi, M. & Taira, K. U6 promoter driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20, 497–500 (2002).
Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J. & Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16, 948–958 (2002).
Paul, C.P., Good, P.D., Winer, I. & Engelke, D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20, 505–508 (2002).
Sui, G. et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99, 5515–5520 (2002).
Yu, J.Y., DeRuiter, S.L. & Turner, D.L. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99, 6047–6052 (2002).
Brummelkamp, T.R., Bernards, R. & Agami, R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2, 243–247 (2002).
McCaffrey, A.P. et al. RNA interference in adult mice. Nature 418, 38–39 (2002).
McManus, M.T. & Sharp, P.A. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3, 737–747 (2002).
Lois, C., Refaeli, Y., Qin, X.F. & Van Parijs, L. Retroviruses as tools to study the immune system. Curr. Opin. Immunol. 13, 496–504 (2001).
Scherr, M. & Eder, M. Gene transfer into hematopoietic stem cells using lentiviral vectors. Curr. Gene Ther. 2, 45–55 (2002).
Tuschl, T. Expanding small RNA interference. Nat. Biotechnol. 20, 446–448 (2002).
Naldini, L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr. Opin. Biotechnol. 9, 457–463 (1998).
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996).
Pfeifer, A., Ikawa, M., Dayn, Y. & Verma, I.M. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc. Natl. Acad. Sci. USA 99, 2140–2145 (2002).
Lois, C., Hong, E.J., Pease, S., Brown, E.J. & Baltimore, D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002).
Hogquist, K.A. et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994).
McManus, M.T. et al. Small interfering RNA-mediated gene silencing in T lymphocytes. J. Immunol. 169, 5754–5760 (2002).
Strasser, A. et al. The role of bim, a proapoptotic BH3-only member of the Bcl-2 family in cell-death control. Ann. NY Acad. Sci. 917, 541–548 (2000).
Willerford, D.M. et al. Interleukin-2 receptor α chain regulates the size and content of the peripheral lymphoid compartment. Immunity 3, 521–530 (1995).
Schmidt, E.V., Christoph, G., Zeller, R. & Leder, P. The cytomegalovirus enhancer: a pan-active control element in transgenic mice. Mol. Cell. Biol. 10, 4406–4411 (1990).
Gertler, F.B., Niebuhr, K., Reinhard, M., Wehland, J. & Soriano, P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87, 227–239 (1996).
Chen, J., Lansford, R., Stewart, V., Young, F. & Alt, F.W. RAG-2–deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc. Natl. Acad. Sci. USA 90, 4528–4532 (1993).
Paddison, P.J. & Hannon, G.J. RNA interference: the new somatic cell genetics? Cancer Cell 2, 17–23 (2002).
Hannon, G.J. RNA interference. Nature 418, 244–251 (2002).
Acknowledgements
We would like to thank J. Bear, J. Chen, P. Sharp, C. Lois and D. Baltimore for their advice and D. Chojnacky, A. Antov, K. Layer, B. Haines, G. Paradis and the Flow Cytometry facility for their technical support. This study was supported by a grant from the David Koch Cancer Research Fund, a Career Development Award from the Arthritis Foundation and the Juvenile Diabetes Foundation (to L.V.P.), a grant from the US National Institutes of Health and a WM Keck Distinguished Young Scholar Award (to F.B.G.). D.A.R. is supported by the Medical Scientist Training Program. C.P.D. is a Howard Hughes Medical Institute Predoctoral Fellow. M.T.M. is a fellow of the Cancer Research Institute. A.V.K. is supported by an Anna Fuller Predoctoral Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Rubinson, D., Dillon, C., Kwiatkowski, A. et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33, 401–406 (2003). https://doi.org/10.1038/ng1117
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1117
This article is cited by
-
HSP27 Modulates Neuropathic Pain by Inhibiting P2X3 Degradation
Molecular Neurobiology (2024)
-
Extracellular vesicles from mesenchymal stem cells reduce neuroinflammation in hippocampus and restore cognitive function in hyperammonemic rats
Journal of Neuroinflammation (2023)
-
A Circular RNA Expressed from the FAT3 Locus Regulates Neural Development
Molecular Neurobiology (2023)
-
Aggressive migration in acidic pH of a glioblastoma cancer stem cell line in vitro is independent of ASIC and KCa3.1 ion channels, but involves phosphoinositide 3-kinase
Pflügers Archiv - European Journal of Physiology (2023)
-
MiR-422a promotes adipogenesis via MeCP2 downregulation in human bone marrow mesenchymal stem cells
Cellular and Molecular Life Sciences (2023)