Abstract
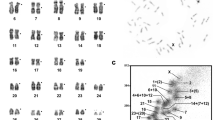
The Syrian cardiomyopathic hamster1 (BIO14.6) has an inherited form of progressive myocardial necrosis and congestive heart failure. Although widely studied2–4 as an animal model for human hypertrophic cardiomyopathy, further genetic analysis has been limited by a scarcity of DMA markers. Until now, only six autosomal linkage groups have been described5 and the number of polymorphic loci was extremely limited. In this study, we applied the restriction landmark genome scanning (RLGS) spot-mapping method6–9 to construct a genetic map of the Syrian hamster (Mesocricetus auratus) using 72 back-cross progeny. Although the polymorphic rate is very low (3–7%) between the strains, 531 polymorphic spots/loci were mapped, showing the power of this approach and reasonable applicability to other organisms lacking a well-defined genetic map. Further, the spot markers which flank the cardiomyopathy (cm) locus were cloned to determine the chromosomal location of cm by fluorescent in situ hybridization (FISH) analysis, resulting in the assignment of the locus to the centromeric region of hamster chromosome 9qa2.1–b1. Several candidate genes responsible for hypertrophic cardiomyopathy in humans have been excluded.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Homburger, F. Baker, J.R., Nixon, C.W. & Whitney, R. Primary,generalized polymyopathy and cardiac necrosis in an inbred line of Syrian hamsters. Med. Exp. 6, 339–345 (1962).
Bajusz, E. & Homburger, F. Myopathies: a meeting report. Science 152, 1112–1113 (1966).
Homburger, F. & Bajusz, E. New models of human disease in Syrian hamsters. J. Am. Med. Assoc. 212, 604–610 (1970).
Wagner, J.A. et al. Calcium antagonist receptors in cardiomyopathic hamster: selective increases in heart, muscle, brain. Science 232, 515–518 (1986).
Robinson, R. Mesocricetus auratus (Syrian Hamster). in Genetic maps: Locus maps of complex genomes 6th edn. (ed. O'Brien, S. J.) (Cold Spring Harbor Laboratory Press, New York, 1993).
Hatada, I. Hayashizaki, Y., Hirotsune, S., Komatsubara, K. & Mukai, T. A genomic scanning method for higher organisms using restriction sites as landmarks on the genome. Proc. Natl. Acad. Sci. USA 88, 9523–9527 (1991).
Hayashizaki, Y. et al. A genetic linkage map of the mouse using restriction landmark genomic scanning (RLGS). Genetics 138, 1207–1238 (1994).
Okuizumi, H. et al. Genetic mapping of restriction landmark genomic scanning (RLGS) loci in the mouse. Electrophoresis 16, 233–240 (1995).
Okazaki, Y. et al. A genetic linkage map of the mouse using an expanded production system of restriction landmark genomic scanning (RLGS Ver.1.8). Biochem. Biophys. Res. Commun. 205, 1922–1929 (1994).
Manly, K.F. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm. Genome 4, 303–313 (1993).
Li, S., Pathak, S. & Hsu, T.C. High resolution G-banding patterns of Syrian hamster chromosomes. Cytogenet. Cell Genet. 33, 295–302 (1982).
Hirotsune, S. et al. Molecular cloning of polymorphic markers on RLGS gel using the spot target cloning method. Biochem. Biophys. Res. Commun. 194, 1406–1412 (1993).
Ohsumi, T. et al. RLGS spot cloning method. Electrophoresis 16, 203–209 (1995).
Matsuda, Y. et al. Location of the mouse complement factor H gene (chf) by FISH analysis and replication R-banding. Cytogenet. Cell Genet. 61, 282–285 (1992).
Roberds, S.L. et al. Disruption of the dystrophin-glycoprotein complex in the cardiomyopathic hamster. J. Biol. Chem. 268, 11496–11499 (1993).
Geisterfer-Lowrance, A.A.T. et al. A molecular basis for familial hypertrophic cardiomyopathy: a β cardiac myosin heavy chain gene missense mutation. Cell 62, 999–1006 (1990).
Thierfelder, L. et al. α-Tropomyosin and cardiacv troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 77, 701–712 (1994).
Okazaki, Y. et.al. Direct detection and isolation of RLGS spot DNA marker tightly linked to a specific trait using RLGS spot-bombing method. Proc. Natl. Acad. Sci. USA 92, 5610–5614 (1995).
Hirotsune, S. et al. The reeler gene encodes a protein with an EGF-like motif expressed by pioneer neurons. Nature Genet. 10, 77–83 (1995).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular cloning: A laboratory manual. 2nd edn. (Cold Spring Harbor Laboratory Press, New York, 1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Okazaki, Y., Okuizumi, H., Ohsumi, T. et al. A genetic linkage map of the Syrian hamster and localization of cardiomyopathy locus on chromosome 9qa2.1–b1 using RLGS spot–mapping. Nat Genet 13, 87–90 (1996). https://doi.org/10.1038/ng0596-87
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng0596-87
This article is cited by
-
Restriction landmark genomic scanning
Nature Protocols (2006)
-
Meeting report: 10th international mouse genome conference
Mammalian Genome (1997)
-
Linkage map of Syrian hamster with restriction landmark genomic scanning
Mammalian Genome (1997)
-
A landmark for orphan genomes?
Nature Genetics (1996)