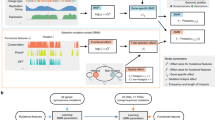
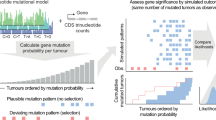
Abstract
Cancer genomics efforts have identified genes and regulatory elements driving cancer development and neoplastic progression. From a microevolution standpoint, these are subject to positive selection. Although elusive in current studies, genes whose wild-type coding sequences are needed for tumor growth are also of key interest. They are expected to experience negative selection and stay intact under pressure of incessant mutation. The detection of significantly mutated (or undermutated) genes is completely confounded by the genomic heterogeneity of cancer mutation1. Here we present a hierarchical framework that allows modeling of coding point mutations. Application of the model to sequencing data from 17 cancer types demonstrates an increased power to detect known cancer driver genes and identifies new significantly mutated genes with highly plausible biological functions. The signal of negative selection is very subtle, but is detectable in several cancer types and in a pan-cancer data set. It is enriched in cell-essential genes identified in a CRISPR screen2, as well as in genes with reported roles in cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Wang, T. et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015).
Polak, P. et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 (2015).
Nik-Zainal, S. et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016).
Rooney, M.S., Shukla, S.A., Wu, C.J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Lindeboom, R.G.H., Supek, F. & Lehner, B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 48, 1112–1118 (2016).
Van den Eynden, J., Basu, S. & Larsson, E. Somatic mutation patterns in hemizygous genomic regions unveil purifying selection during tumor evolution. PLoS Genet. 12, e1006506 (2016).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Preprint at bioRxivhttp://dx.doi.org/10.1101/132324 (2017).
McFarland, C.D., Korolev, K.S., Kryukov, G.V., Sunyaev, S.R. & Mirny, L.A. Impact of deleterious passenger mutations on cancer progression. Proc. Natl. Acad. Sci. USA 110, 2910–2915 (2013).
Melton, C., Reuter, J.A., Spacek, D.V. & Snyder, M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 47, 710–716 (2015).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Scharenberg, M.A., Chiquet-Ehrismann, R. & Asparuhova, M.B. Megakaryoblastic leukemia protein-1 (MKL1): increasing evidence for an involvement in cancer progression and metastasis. Int. J. Biochem. Cell Biol. 42, 1911–1914 (2010).
Sheriff, S. et al. Neuropeptide Y Y5 receptor promotes cell growth through extracellular signal–regulated kinase signaling and cyclic AMP inhibition in a human breast cancer cell line. Mol. Cancer Res. 8, 604–614 (2010).
Law, M.H. et al. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet. 47, 987–995 (2015).
Nürnberg, A., Kitzing, T. & Grosse, R. Nucleating actin for invasion. Nat. Rev. Cancer 11, 177–187 (2011).
Grabarczyk, P. et al. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene 26, 3797–3810 (2007).
Adams, J.M. & Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26, 1324–1337 (2007).
Fine, B. et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science 325, 1261–1265 (2009).
Boggs, A.E. et al. α-Tubulin acetylation elevated in metastatic and basal-like breast cancer cells promotes microtentacle formation, adhesion, and invasive migration. Cancer Res. 75, 203–215 (2015).
Bilbe, G. et al. Restin: a novel intermediate filament–associated protein highly expressed in the Reed–Sternberg cells of Hodgkin's disease. EMBO J. 11, 2103–2113 (1992).
Park, J.H. et al. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 70, 2759–2769 (2010).
Fielding, A.B., Lim, S., Montgomery, K., Dobreva, I. & Dedhar, S. A critical role of integrin-linked kinase, ch-TOG and TACC3 in centrosome clustering in cancer cells. Oncogene 30, 521–534 (2011).
Xie, K., Doles, J., Hemann, M.T. & Walker, G.C. Error-prone translesion synthesis mediates acquired chemoresistance. Proc. Natl. Acad. Sci. USA 107, 20792–20797 (2010).
Futreal, P.A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Binamé, F. et al. Cancer-associated mutations in the protrusion-targeting region of p190RhoGAP impact tumor cell migration. J. Cell Biol. 214, 859–873 (2016).
Baud, V. & Karin, M. Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 8, 33–40 (2009).
Pasquale, E.B. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180 (2010).
Tanaka, I. et al. LIM-domain protein AJUBA suppresses malignant mesothelioma cell proliferation via Hippo signaling cascade. Oncogene 34, 73–83 (2015).
Ullah, F. et al. Promoter methylation status modulate the expression of tumor suppressor (RbL2/p130) gene in breast cancer. PLoS One 10, e0134687 (2015).
Felsenstein, J. The evolutionary advantage of recombination. Genetics 78, 737–756 (1974).
Good, B.H. & Desai, M.M. Deleterious passengers in adapting populations. Genetics 198, 1183–1208 (2014).
Supek, F., Miñana, B., Valcárcel, J., Gabaldón, T. & Lehner, B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 (2014).
Lawrence, M.S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Meyer, M.J. et al. mutation3D: cancer gene prediction through atomic clustering of coding variants in the structural proteome. Hum. Mutat. 37, 447–456 (2016).
Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank S. Schiffels and L. Mirny for helpful discussion during the conception of the project and feedback on the manuscript, as well as P. Polak and Z. Li for critical reading of the manuscript. We also thank V. Seplyarskiy for advice on mutational processes and I. Adzhubey for support with the setup of the web interface for CBaSE. This work was supported by US National Institutes of Health (NIH) grants U54 CA143874, R01 MH101244, and R01 GM078598 (S.S.).
Author information
Authors and Affiliations
Contributions
D.W. designed the statistical framework, wrote code, analyzed and interpreted data, created the web interface, and wrote the manuscript. S.S. supervised the project, gave technical and conceptual advice, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–31 and Supplementary Note (PDF 8815 kb)
Supplementary Tables 1–16
Supplementary Tables 1–16 (XLSX 35583 kb)
Rights and permissions
About this article
Cite this article
Weghorn, D., Sunyaev, S. Bayesian inference of negative and positive selection in human cancers. Nat Genet 49, 1785–1788 (2017). https://doi.org/10.1038/ng.3987
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3987
This article is cited by
-
Cancer genomes tolerate deleterious coding mutations through somatic copy number amplifications of wild-type regions
Nature Communications (2023)
-
Centuries of genome instability and evolution in soft-shell clam, Mya arenaria, bivalve transmissible neoplasia
Nature Cancer (2023)
-
MHC II immunogenicity shapes the neoepitope landscape in human tumors
Nature Genetics (2023)
-
Unraveling the Drivers of Tumorigenesis in the Context of Evolution: Theoretical Models and Bioinformatics Tools
Journal of Molecular Evolution (2023)
-
The impact of rare germline variants on human somatic mutation processes
Nature Communications (2022)