Abstract
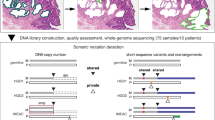
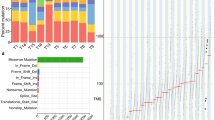
The molecular genetic relationship between esophageal adenocarcinoma (EAC) and its precursor lesion, Barrett's esophagus, is poorly understood. Using whole-genome sequencing on 23 paired Barrett's esophagus and EAC samples, together with one in-depth Barrett's esophagus case study sampled over time and space, we have provided the following new insights: (i) Barrett's esophagus is polyclonal and highly mutated even in the absence of dysplasia; (ii) when cancer develops, copy number increases and heterogeneity persists such that the spectrum of mutations often shows surprisingly little overlap between EAC and adjacent Barrett's esophagus; and (iii) despite differences in specific coding mutations, the mutational context suggests a common causative insult underlying these two conditions. From a clinical perspective, the histopathological assessment of dysplasia appears to be a poor reflection of the molecular disarray within the Barrett's epithelium, and a molecular Cytosponge technique overcomes sampling bias and has the capacity to reflect the entire clonal architecture.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012).
Esserman, L.J. et al. Addressing overdiagnosis and overtreatment in cancer: a prescription for change. Lancet Oncol. 15, e234–e242 (2014).
Desai, T.K. et al. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett's oesophagus: a meta-analysis. Gut 61, 970–976 (2012).
Corley, D.A. et al. Impact of endoscopic surveillance on mortality from Barrett's esophagus–associated esophageal adenocarcinomas. Gastroenterology 145, 312–319 (2013).
Shaheen, N.J. & Hur, C. Garlic, silver bullets, and surveillance upper endoscopy for Barrett's esophagus. Gastroenterology 145, 273–276 (2013).
Leedham, S.J. et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett's oesophagus. Gut 57, 1041–1048 (2008).
Maley, C.C. et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett's esophagus. Cancer Res. 64, 3414–3427 (2004).
Schulmann, K. et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett's-associated neoplastic progression and predicts progression risk. Oncogene 24, 4138–4148 (2005).
Reid, B.J. et al. Predictors of progression in Barrett's esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am. J. Gastroenterol. 96, 2839–2848 (2001).
Kastelein, F. et al. Aberrant p53 protein expression is associated with an increased risk of neoplastic progression in patients with Barrett's oesophagus. Gut 62, 1676–1683 (2013).
Maley, C.C. Multistage carcinogenesis in Barrett's esophagus. Cancer Lett. 245, 22–32 (2007).
Fitzgerald, R.C. Dissecting out the genetic origins of Barrett's oesophagus. Gut 57, 1033–1034 (2008).
Weaver, J.M. et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat. Genet. 46, 837–843 (2014).
Agrawal, N. et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2, 899–905 (2012).
Chong, I.Y. et al. The genomic landscape of oesophagogastric junctional adenocarcinoma. J. Pathol. 231, 301–310 (2013).
Dulak, A.M. et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 45, 478–486 (2013).
Alexandrov, L.B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Chapman, M.A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Ellis, M.J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012).
Kan, Z. et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 23, 1422–1433 (2013).
Bass, A.J. et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 43, 964–968 (2011).
Dulak, A.M. et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 72, 4383–4393 (2012).
Schneider, P.M. et al. Mutations of p53 in Barrett's esophagus and Barrett's cancer: a prospective study of ninety-eight cases. J. Thorac. Cardiovasc. Surg. 111, 323–331 discussion 331–333 (1996).
Dolan, K., Walker, S.J., Gosney, J., Field, J.K. & Sutton, R. TP53 mutations in malignant and premalignant Barrett's esophagus. Dis. Esophagus 16, 83–89 (2003).
Li, X. et al. Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett's esophagus. Cancer Prev. Res. (Phila.) 7, 114–127 (2014).
Orman, E.S., Li, N. & Shaheen, N.J. Efficacy and durability of radiofrequency ablation for Barrett's Esophagus: systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 11, 1245–1255 (2013).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Raczy, C. et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 29, 2041–2043 (2013).
Saunders, C.T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
Gusnanto, A., Wood, H.M., Pawitan, Y., Rabbitts, P. & Berri, S. Correcting for cancer genome size and tumour cell content enables better estimation of copy number alterations from next-generation sequence data. Bioinformatics 28, 40–47 (2012).
Acknowledgements
We thank the Human Research Tissue Bank, which is supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. This study was partly funded by a project grant from Cancer Research UK. R.C.F. has programmatic funding from the Medical Research Council and infrastructure support from the Biomedical Research Centre and the Experimental Medicine Centre. We would like to thank all the patients who took part in the study. We thank M. Dunning for his bioinformatics assistance. We thank the Edinburgh Experimental Cancer Medicine Centre.
Author information
Authors and Affiliations
Consortia
Contributions
R.C.F. conceived the overall study and takes responsibility for the data integrity. C.S.R.-I., J.B. and R.K.C. analyzed the data. C.S.R.-I. extracted the samples for patient AHM1051. A.W. developed the targeted sequencing data visualization tool. C.S.R.-I., J.B., R.K.C., H.N., J.M.J.W., M.R., S.H., D.B. and R.C.F. designed various aspects of the study. H.N. performed the TruSeq Custom Amplicon (TSCA) assay. C.S.R.-I., J.B. and A.G.L. performed the statistical analysis. M.d.P. collected endoscopic samples for patient AHM1051. M.O'D. and S.M. performed the histopathological diagnosis. S.I. developed the copy number pipeline, and R.K.C. and M.H. performed the copy number analysis. Z.K. ran the whole-genome sequencing of patient AHM1051. R.C.F., S.H., D.B. and M.R. supervised the study. C.S.R.-I., J.B., R.K.C. and R.C.F. wrote the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.C.F. developed the Cytosponge technology, which has been licensed by MRC-Technology to Covidien. R.C.F. has no direct pecuniary interest. J.B., A.W., R.K.C., H.N., S.I., M.H., Z.K., M.R., S.H. and D.B. are employees of Illumina.
Integrated supplementary information
Supplementary Figure 1 Multiple samples from five patients (P4, P8, P14, P15 and P17) with Barrett's esophagus and adjacent EAC show that the poor overlap is not a result of sampling bias.
Bar graphs showing the number of SNVs for samples of Barrett's esophagus (BE) and cancer (C) that are either present in all samples (teal), present in all BE samples and absent in all cancer samples (pink), or present in all cancer samples and absent in all BE samples (gray) for each particular patient. Purple bars show the number of remaining variants not accounted for in the other groups (for example, present in a proportion of samples).
Supplementary Figure 2 EAC samples display multiple copy number aberrations compared to paired Barrett's samples.
(a) Copy number plots showing two examples of Barrett's esophagus genomes that are majority copy number 2, compared to the matched EAC samples, which contain multiple gains and losses. (b) Similar plots but for patients where copy number events can be seen in the Barrett's esophagus sample as well as in the EAC sample (but are not significant enough to make a call). (c) Examples of copy number events in the Barrett's esophagus sample that are not seen in the matching EAC sample. The top track shows the depth ratio in a 20-kb window, and the bottom track shows the B-allele ratio.
Supplementary Figure 3 Principal-component analysis of mutational context.
The counts of each different mutational context in each of the three sets of SNVs (common, Barrett's esophagus (BE) unique, EAC unique) were used as input for a principal-component analysis. There is some separation between the common SNVs and the unique SNVs in the second component, but the differences between the groups are not striking (6.9% variance in the second component). (b) Plot showing the weights of the 96 different mutational contexts. Four mutational contexts—C(A>C)C, G(C>G)G and C(A>T)C (all at the top right-hand side of the plot) as well as T(C>G)A (bottom right)—seem to be the main cause for the separation in the second component.
Supplementary Figure 4 Copy number plot for one of the Barrett's esophagus samples with low-grade dysplasia.
Copy number plot for one of the Barrett's esophagus samples with low-grade dysplasia that was sent for whole-genome sequencing. Large deletions on multiple chromosomes, including chromosomes 5, 11, 13, 18 and 21, can be seen.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Tables 1–4. (PDF 1340 kb)
Supplementary Data Set
Nebula: custom-made data browser to investigate and visualize the results from the 1,443 targets that were sequenced for 73 Barrett's esophagus samples from patient AHM1051. (ZIP 7637 kb)
Rights and permissions
About this article
Cite this article
Ross-Innes, C., Becq, J., Warren, A. et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet 47, 1038–1046 (2015). https://doi.org/10.1038/ng.3357
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3357
This article is cited by
-
Evolving copy number gains promote tumor expansion and bolster mutational diversification
Nature Communications (2024)
-
Integrated clinical and genomic analysis identifies driver events and molecular evolution of colitis-associated cancers
Nature Communications (2023)
-
Mutational signature dynamics shaping the evolution of oesophageal adenocarcinoma
Nature Communications (2023)
-
Targeted genetic and epigenetic profiling of esophageal adenocarcinomas and non-dysplastic Barrett’s esophagus
Clinical Epigenetics (2022)
-
Utility of ancillary studies in the diagnosis and risk assessment of Barrett's esophagus and dysplasia
Modern Pathology (2022)