Abstract
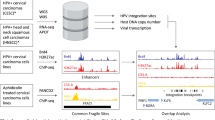
Human papillomavirus (HPV) integration is a key genetic event in cervical carcinogenesis1. By conducting whole-genome sequencing and high-throughput viral integration detection, we identified 3,667 HPV integration breakpoints in 26 cervical intraepithelial neoplasias, 104 cervical carcinomas and five cell lines. Beyond recalculating frequencies for the previously reported frequent integration sites POU5F1B (9.7%), FHIT (8.7%), KLF12 (7.8%), KLF5 (6.8%), LRP1B (5.8%) and LEPREL1 (4.9%), we discovered new hot spots HMGA2 (7.8%), DLG2 (4.9%) and SEMA3D (4.9%). Protein expression from FHIT and LRP1B was downregulated when HPV integrated in their introns. Protein expression from MYC and HMGA2 was elevated when HPV integrated into flanking regions. Moreover, microhomologous sequence between the human and HPV genomes was significantly enriched near integration breakpoints, indicating that fusion between viral and human DNA may have occurred by microhomology-mediated DNA repair pathways2. Our data provide insights into HPV integration-driven cervical carcinogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Referenced accessions
NCBI Reference Sequence
References
Pett, M. & Coleman, N. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J. Pathol. 212, 356–367 (2007).
Lawson, A.R. et al. RAF gene fusion breakpoints in pediatric brain tumors are characterized by significant enrichment of sequence microhomology. Genome Res. 21, 505–514 (2011).
zur Hausen, H. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2, 342–350 (2002).
Crosbie, E.J., Einstein, M.H., Franceschi, S. & Kitchener, H.C. Human papillomavirus and cervical cancer. Lancet 382, 889–899 (2013).
Shi, Y. et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat. Genet. 45, 918–922 (2013).
Stanley, M.A., Pett, M.R. & Coleman, N. HPV: from infection to cancer. Biochem. Soc. Trans. 35, 1456–1460 (2007).
Wentzensen, N., Vinokurova, S. & von Knebel Doeberitz, M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 64, 3878–3884 (2004).
Hudelist, G. et al. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol. Oncol. 92, 873–880 (2004).
Arias-Pulido, H., Peyton, C.L., Joste, N.E., Vargas, H. & Wheeler, C.M. Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J. Clin. Microbiol. 44, 1755–1762 (2006).
el Awady, M.K., Kaplan, J.B., O'Brien, S.J. & Burk, R.D. Molecular analysis of integrated human papillomavirus 16 sequences in the cervical cancer cell line SiHa. Virology 159, 389–398 (1987).
Thorland, E.C. et al. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 60, 5916–5921 (2000).
Luft, F. et al. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int. J. Cancer 92, 9–17 (2001).
Kalantari, M., Blennow, E., Hagmar, B. & Johansson, B. Physical state of HPV16 and chromosomal mapping of the integrated form in cervical carcinomas. Diagn. Mol. Pathol. 10, 46–54 (2001).
Xu, B. et al. Multiplex identification of human papillomavirus 16 DNA integration sites in cervical carcinomas. PLoS ONE 8, e66693 (2013).
Klaes, R. et al. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 59, 6132–6136 (1999).
Li, W. et al. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics 102, 338–344 (2013).
Dall, K.L. et al. Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res. 68, 8249–8259 (2008).
Ferber, M.J. et al. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene 22, 7233–7242 (2003).
Schmitz, M. et al. Loss of gene function as a consequence of human papillomavirus DNA integration. Int. J. Cancer 131, E593–E602 (2012).
Peter, M. et al. MYC activation associated with the integration of HPV DNA at the MYC locus in genital tumors. Oncogene 25, 5985–5993 (2006).
Boulet, G.A. et al. Human papillomavirus 16 load and E2/E6 ratio in HPV16-positive women: biomarkers for cervical intraepithelial neoplasia >or=2 in a liquid-based cytology setting? Cancer Epidemiol. Biomarkers Prev. 18, 2992–2999 (2009).
Gradíssimo Oliveira, A., Delgado, C., Verdasca, N. & Pista, A. Prognostic value of human papillomavirus types 16 and 18 DNA physical status in cervical intraepithelial neoplasia. Clin. Microbiol. Infect. 19, E447–E450 (2013).
Adey, A. et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 500, 207–211 (2013).
Ojesina, A.I. et al. Landscape of genomic alterations in cervical carcinomas. Nature 506, 371–375 (2014).
Schmitz, M., Driesch, C., Jansen, L., Runnebaum, I.B. & Durst, M. Non-random integration of the HPV genome in cervical cancer. PLoS ONE 7, e39632 (2012).
Lee, J.A., Carvalho, C.M. & Lupski, J.R.A. DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (2007).
Zhang, F. et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 41, 849–853 (2009).
Verdin, H. et al. Microhomology-mediated mechanisms underlie non-recurrent disease-causing microdeletions of the FOXL2 gene or its regulatory domain. PLoS Genet. 9, e1003358 (2013).
Campitelli, M. et al. Human papillomavirus mutational insertion: specific marker of circulating tumor DNA in cervical cancer patients. PLoS ONE 7, e43393 (2012).
Das, P. et al. HPV genotyping and site of viral integration in cervical cancers in Indian women. PLoS ONE 7, e41012 (2012).
Karlsen, F. et al. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J. Clin. Microbiol. 34, 2095–2100 (1996).
Baay, M.F. et al. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J. Clin. Microbiol. 34, 745–747 (1996).
Walboomers, J.M. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19 (1999).
Woodman, C.B., Collins, S.I. & Young, L.S. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer 7, 11–22 (2007).
Stanley, M.A., Browne, H.M., Appleby, M. & Minson, A.C. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int. J. Cancer 43, 672–676 (1989).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Lettice, L.A. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 (2003).
Li, L. et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 122, 902–911 (2013).
Sung, W.K. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 44, 765–769 (2012).
Burk, R.D., Harari, A. & Chen, Z. Human papillomavirus genome variants. Virology 445, 232–243 (2013).
Cer, R.Z. et al. Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools. Nucleic Acids Res. 41, D94–D100 (2013).
Abeysinghe, S.S., Chuzhanova, N., Krawczak, M., Ball, E.V. & Cooper, D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum. Mutat. 22, 229–244 (2003).
Acknowledgements
This work was supported by funds from the National Development Program (973) for the Key Basic Research of China (2015CB553903 and 2013CB911304) and the National Natural Science Funding of China (81230038, 81372805, 81172466, 81272859, 81372801, 81372804, 81402158, 81370469, 81372806, 81370469, 81172468, 81101964, 81230052 and 81302266). The study is also sponsored by the Chinese 863 Program (2012AA02A507, 2012AA02A201), Guangdong Enterprise Key Laboratory of Human Disease Genomics, ShenZhen Engineering Laboratory for Clinical Molecular Diagnostic and China National GeneBank–Shenzhen. We thank all participants recruited for this study. We thank our colleagues from BGI, H. Huang, L. Huang, X. Zhuang, L. Lin, H. Cao, X. Fang, X. Zhang, Y. Shuang and H. Yang for sequencing and analysis.
Author information
Authors and Affiliations
Contributions
D.M. took full responsibility for the study, especially in conceiving, designing and supervising the research together with Z.H. and H.W. W.W., J.Z., J.W. and X.X. participated in the study design. G.C., Q.G., Shuang Li., L.X., C.W., S. Liao, X.M., P.W., K.L. and S.W. supervised the diagnosis of patients and subject recruitment. D.Z., H.S., W.J., W.L., X.Z., H.L., X.F., Shuaicheng Li and X.L. performed statistical analysis. W.L., X.Z. and Y.Z. performed HIVID and RNA-seq analysis. W.J. performed FuseSV (in-house software for MH analysis) and WGS analysis. W.D., L.Y., L.W., H.S., X.W., C.Z., W.C., T.T., A.F. and Z.W. performed the experiments. The manuscript was drafted by Z.H., D.Z., W.J., W.L. and X.Z. under the supervision of D.M., H.W. and X.X. All authors critically reviewed the article and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–29 and Supplementary Note (PDF 23284 kb)
Supplementary Tables 1–28
Supplementary Tables 1–28 (XLSX 3257 kb)
Source data
Rights and permissions
About this article
Cite this article
Hu, Z., Zhu, D., Wang, W. et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet 47, 158–163 (2015). https://doi.org/10.1038/ng.3178
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3178
This article is cited by
-
Intratumoural microbiota: a new frontier in cancer development and therapy
Signal Transduction and Targeted Therapy (2024)
-
Elucidating the clonal relationship of esophageal second primary tumors in patients with laryngeal squamous cell carcinoma
Infectious Agents and Cancer (2023)
-
The analysis of HPV integration sites based on nanopore sequencing and the profiling changes along the course of photodynamic therapy
BMC Cancer (2023)
-
Upregulation of FAM83F by c-Myc promotes cervical cancer growth and aerobic glycolysis via Wnt/β-catenin signaling activation
Cell Death & Disease (2023)
-
Carcinogenesis and management of human papillomavirus-associated cervical cancer
International Journal of Clinical Oncology (2023)