Abstract
Febrile seizures affect 2–4% of all children1 and have a strong genetic component2. Recurrent mutations in three main genes (SCN1A, SCN1B and GABRG2)3,4,5 have been identified that cause febrile seizures with or without epilepsy. Here we report the identification of mutations in STX1B, encoding syntaxin-1B6, that are associated with both febrile seizures and epilepsy. Whole-exome sequencing in independent large pedigrees7,8 identified cosegregating STX1B mutations predicted to cause an early truncation or an in-frame insertion or deletion. Three additional nonsense or missense mutations and a de novo microdeletion encompassing STX1B were then identified in 449 familial or sporadic cases. Video and local field potential analyses of zebrafish larvae with antisense knockdown of stx1b showed seizure-like behavior and epileptiform discharges that were highly sensitive to increased temperature. Wild-type human syntaxin-1B but not a mutated protein rescued the effects of stx1b knockdown in zebrafish. Our results thus implicate STX1B and the presynaptic release machinery in fever-associated epilepsy syndromes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
Ensembl
NCBI Reference Sequence
Change history
07 November 2014
In the version of this article initially published online, the amplitudes of the multispike bursts associated with epileptiform paroxysmal events were incorrectly reported as being equal to baseline or exceeding it by threefold. The amplitudes should have been reported as exceeding baseline by at least threefold. The error has been corrected for the print, PDF and HTML versions of this article.
09 December 2014
In the version of this article initially published, the name of author Frank Lehmann-Horn was misspelled. The error has been corrected in the HTML and PDF versions of the article.
References
Berg, A.T., Jallon, P. & Preux, P.M. The epidemiology of seizure disorders in infancy and childhood: definitions and classifications. Handb. Clin. Neurol. 111, 391–398 (2013).
Eckhaus, J. et al. Genetics of febrile seizure subtypes and syndromes: a twin study. Epilepsy Res. 105, 103–109 (2013).
Escayg, A. et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat. Genet. 24, 343–345 (2000).
Wallace, R.H. et al. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel β1 subunit gene SCN1B. Nat. Genet. 19, 366–370 (1998).
Wallace, R.H. et al. Mutant GABA(A) receptor γ2-subunit in childhood absence epilepsy and febrile seizures. Nat. Genet. 28, 49–52 (2001).
Südhof, T.C. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690 (2013).
Lerche, H. et al. Generalized epilepsy with febrile seizures plus: further heterogeneity in a large family. Neurology 57, 1191–1198 (2001).
Weber, Y.G. et al. A BFIS-like syndrome with late onset and febrile seizures: suggestive linkage to chromosome 16p11.2-16q12.1. Epilepsia 49, 1959–1964 (2008).
Chen, W.J. et al. Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat. Genet. 43, 1252–1255 (2011).
Wang, J.L. et al. Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain 134, 3493–3501 (2011).
Lee, H.Y. et al. Mutations in the gene PRRT2 cause praroxysmal kinesigenic dyskinesia with infantile convulsions. Cell Rep. 1, 2–12 (2012).
Schubert, J. et al. PRRT2 mutations are the major cause of benign familial infantile seizures. Hum. Mutat. 33, 1439–1443 (2012).
Heron, S.E. et al. PRRT2 mutations cause benign familial infantile epilepsy and infantile convulsions with choreoathetosis syndrome. Am. J. Hum. Genet. 90, 152–160 (2012).
Becker, F. et al. PRRT2-related disorders: further PKD and ICCA cases and review of the literature. J. Neurol. 260, 1234–1244 (2013).
Saitsu, H. et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 40, 782–788 (2008).
Lemke, J.R. et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia 53, 1387–1398 (2012).
Nasevicius, A. & Ekker, S.C. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 26, 216–220 (2000).
Afrikanova, T. et al. Validation of the zebrafish pentylenetetrazol seizure model: locomotor versus electrographic responses to antiepileptic drugs. PLoS ONE 8, e54166 (2013).
Jefferys, J.G. et al. Mechanisms of physiological and epileptic HFO generation. Prog. Neurobiol. 98, 250–264 (2012).
Epi4K Consortium & Epilepsy Phenome/Genome Project. De novo mutations in epileptic encephalopathies. Nature 501, 217–221 (2013).
Bragina, L., Giovedì, S., Barbaresi, P., Benfenati, F. & Conti, F. Heterogeneity of glutamatergic and GABAergic release machinery in cerebral cortex: analysis of synaptogyrin, vesicle-associated membrane protein, and syntaxin. Neuroscience 165, 934–943 (2010).
Pietri, T., Manalo, E., Ryan, J., Saint-Amant, L. & Washbourne, P. Glutamate drives the touch response through a rostral loop in the spinal cord of zebrafish embryos. Dev. Neurobiol. 69, 780–795 (2009).
Zhou, P. et al. Syntaxin-1 N-peptide and Habc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 32, 159–171 (2013).
Gerber, S.H. et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science 321, 1507–1510 (2008).
Foletti, D.L., Lin, R., Finley, M.A. & Scheller, R.H. Phosphorylated syntaxin 1 is localized to discrete domains along a subset of axons. J. Neurosci. 20, 4535–4544 (2000).
Rüschendorf, F. & Nürnberg, P. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics 21, 2123–2125 (2005).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. GRR: graphical representation of relationship errors. Bioinformatics 17, 742–743 (2001).
O'Connell, J.R. & Weeks, D.E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63, 259–266 (1998).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Sobel, E. & Lange, K. Descent graphs in pedigree analysis: applications to haplotyping,location scores, and marker sharing statistics. Am. J. Hum. Genet. 58, 1323–1337 (1996).
Sobel, E., Sengul, H. & Weeks, D.E. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum. Hered. 52, 121–131 (2001).
Mukhopadhyay, N., Almasy, L., Schroeder, M., Mulvihill, W.P. & Weeks, D.E. Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics 21, 2556–2557 (2005).
Thiele, H. & Nürnberg, P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics 21, 1730–1732 (2005).
Purcell, S. et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 81, 559–575 (2007).
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Albers, C.A. et al. Dindel: accurate indel calls from short-read data. Genome Res. 21, 961–973 (2011).
Yeo, G. & Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 (2004).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling nanoarrays. Science 327, 78–81 (2010).
Roach, J.C. et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328, 636–639 (2010).
Roach, J.C. et al. Chromosomal haplotypes by genetics phasing of human families. Am. J. Hum. Genet. 89, 382–397 (2011).
Summerton, J. & Weller, D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7, 187–195 (1997).
Acknowledgements
We thank the families and their relatives for their cooperation. We thank P. Verstreken (VIB, KU Leuven) for providing access to the electrophysiology equipment. This study was supported by the Federal Ministry for Education and Research (BMBF; NGFNplus/EMINet to H.L., P.N. and T.S., 01GS08123 and 01GS08120; IonNeurONet to H.L., K.J.-R., F.L.-H. and S. Biskup, 01GM1105), funding to the EuroEPINOMICS Consortium coordinated by the European Science Foundation from the German Research Foundation (DFG; Le1030/11-1 to H.L., He5415/3-1 to I.H., Sa434/5-1 to T.S. and Nu50/8-1 to P.N.), the Swiss National Fund (32EP30_136042/1 to J.R.L.) and the Fund for Scientific Research Flanders (GA13611N to P.D.J.). Y.G.W. and I.H. were funded by the German Research Foundation (DFG: WE4896/3-1 and HE5415/6-1). Y.G.W. and S. Biskup received intramural funding from the University of Tübingen (AKF 297-0-0). The German Society for Epileptology (DGfE) gave funding to H.L., Y.G.W., I.H. and H.M., and the foundation no-epilep gave funding to H.L. I.H. and H.M. received further support through intramural grants from the University of Kiel and infrastructural support from the Institute of Clinical Molecular Biology in Kiel, supported in part by the popgen 2.0 network (P2N) through a grant from the BMBF (01EY1103) and Cluster of Excellence 'Inflammation at Interfaces' and 'Future Ocean' programs funded by the DFG. The study was further supported by a Center for Systems Biology P50 grant (GM076547, US National Institutes of Health) to L.H. and by the University of Luxembourg–Institute for Systems Biology Program (P.M., J.C.R., G.G., L.H. and D.J.G.). M. Brunkow provided project management for the whole-genome sequencing. P.M. was supported by 'Le Plan Technologies de la Santé par le Gouvernment du Grand-Duché de Luxembourg' through the Luxembourg Centre for Systems Biomedicine (LCSB), University of Luxembourg. A. Kecskés and K.H. are recipients of a PhD fellowship from the Agency for Innovation by Science and Technology (IWT, Flanders). A. Suls is a postdoctoral fellow of the Research Foundation–Flanders (FWO Vlaanderen). M.L. and M.J. were supported by the LCSB KT program. F.L.-H. and K.J.-R. are supported by the Hertie Foundation. S. Baulac received financial support from the program 'Investissements d'Avenir' (ANR-10-IAIHU-06).
Author information
Authors and Affiliations
Consortia
Contributions
H.L., Y.G.W., C.V.E. and A.D.C. designed the study. F.B., H.M., J.R.L., C.G.F.d.K., I.N., B.K., S.W., T.D., E.H.B., S. Baulac, R.S.M., H.H., B.P.C.K., P.D.J., I.H., Y.G.W., H.L. and the EuroEPINOMICS RES Consortium recruited and phenotyped subjects. Y.G.W., T.S., F.R., K.J.-R. and F.L.-H. performed or supervised whole-genome linkage analysis. J.S., P.M., A. Suls, J.R.L., H.T., K.K., A. Kawalia, E.R., J.C.R., G.G., L.H., D.J.G., S. Biskup, P.D.J., I.H., R.B., P.N., Y.G.W. and H.L. performed or supervised whole-exome, whole-genome or targeted panel sequencing. J.S. and M.R.T. performed segregation analysis and Sanger validation. J.S. and K.H. performed cohort screening by Sanger sequencing. A.C. and I.N. performed FISH and array CGH experiments. A. Siekierska, A. Kecskés and M.J. performed zebrafish knockdown studies. M.L., A. Siekierska, C.H. and B.M. performed zebrafish LFP and spectral analysis. P.A.M.d.W., A.D.C. and C.V.E. supervised all zebrafish studies. J.S., A. Siekierska, M.L., P.M., F.B., H.M., J.R.L., T.S., I.H., A.D.C., C.V.E., Y.G.W. and H.L. wrote the manuscript. All authors revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
P.N. is a founder, CEO and shareholder of ATLAS Biolabs. ATLAS Biolabs is a service provider for genomic analyses. A.D.C. and R.B. are shareholders of Theracule.
Additional information
A full list of members and affiliations appears in the Supplementary Note.
Integrated supplementary information
Supplementary Figure 1 F1 pedigree as used for the whole-genome linkage analysis.
Family 1 exhibits very high phenotypic variability. As described in the main text, we therefore defined six core members with the most consistent phenotype (407 = IV.5, 408 = IV.6, 411 = IV.40, 502 = V.4, 503 = V.5, 504 = V.8) ( Fig. 1a , Supplementary Table 6 and Supplementary Note ; the phenotypes are described in further detail in ref. 7 of the main text). Linkage analysis was performed by considering only the six core members and all obligate carriers of the trait-related mutation to be affected (black symbols), whereas all others with febrile seizures, epilepsy or seizure-suggestive symptoms were given a status of unknown affectedness (gray symbols). Individuals with no history of febrile or afebrile seizures are indicated with white symbols. Male individuals are represented by squares, and female individuals are represented by circles. Mode of inheritance: autosomal dominant with 90% penetrance; mutation frequency of 0.001; phenocopy rate: 1%.
Supplementary Figure 2 Genome-wide linkage results.
The figure shows the result of the genome-wide linkage analysis. The most significant parametric LOD score of 4.27 was detected on chromosome 16p11.2. This region harbors the STX1B gene.
.
Supplementary Figure 3 Parametric multipoint linkage results in the chromosomal region 16p11.2.
The figure shows the region 16p11.2 in detail. The linkage region starts at 29,698,606 and ends at 48,890,475 spanning a 19-Mb risk haplotype at 16p11.2. STX1B starts at 31,000,576 and ends at 31,021,828 and is located in this region.
Supplementary Figure 4 Segregation of the risk haplotype at the 16p11.2 locus in family F1.
The black rectangle indicates the location of the 19-Mb risk haplotype. This region is flanked by the SNPs rs4563056 (chr. 16: 29,698,606, hg19) and rs12598215 (chr. 16: 48,890,475, hg19). The risk haplotype at 16p11.2 is shared by all family members with febrile or afebrile plus febrile seizures. Two family members with IGE syndromes with afebrile generalized seizures only and two family members with an ambiguous history of seizures did not carry the risk haplotype, suggesting intrafamilial locus heterogeneity with the involvement of additional genetic risk factors. The identified STX1B truncation mutation (c.166C>T; p.Gln56*) displays perfect cosegregation with the risk haplotype and likely represents the causative mutation in family F1.
Supplementary Figure 5 Analysis pipeline for F1-specific genetic variants using whole-genome sequencing (WGS).
WGS was performed in seven family members, including six core individuals with febrile and afebrile seizures (IV.5, IV.6, IV.40, V.4, V.5, V.) and one individual with classical childhood absence epilepsy without febrile seizures (IV.19), as described in the main text. Rare and deleterious variants that (i) were shared by the six core members alone and (ii) shared by the core members and IV.19 were identified using genome annotation, public databases and inheritance state analysis (Roach et al., 2010). Complete Genomics (CG) Analysis tools (cgatools) were used to build the union of all variants and to test for the presence of variants in individual genomes. ISCA32 is used to identify haplotype blocks shared by genomes. Ingenuity Variant Analysis is used to annotate and filter for rare and deleterious variants. CMS is used to filter out likely false positive variants.
Supplementary Figure 6 Shared haplotype blocks
Each chromosome in this schematic karyotype is used to represent information abstracted from the shared chromosomal regions (dark blue). It is vertically split to indicate regions shared by the six genomes of affected core members presenting with febrile and afebrile seizures (left) and these same six genomes plus one genome for IV.19 presenting with classical childhood absence epilepsy without febrile seizures (right). The candidate variant in STX1B (depicted in red) lies in a region on chromosome 16 shared only by the genomes of the six core members.
Supplementary Figure 7 De novo microdeletion in an MAE patient (family F6).
The top red bar represents the 0.8-Mb deleted region. STX1B is located in this region. Other genes in this region are listed below. In the lower part of the figure, known segmental duplications (blue) and deletions (red) are shown. The figure was generated using the Database of Genomic Variation website (http://dgv.tcag.ca/dgv/app/home). Targets were mapped to hg19.
Supplementary Figure 8 Sanger sequences of detected missense, nonsense and indel variants.
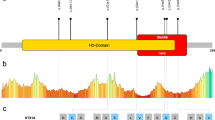
The figure shows the Sanger sequencing chromatograms for the five detected mutations in five independent families. Labels from A to E correspond to the labels in Figure 1. The upper nucleotide sequence shows the reference sequence (ENST00000215095). The chromatogram below shows the wild-type (WT) chromatogram and corresponding sequence. The second chromatogram shows the heterozygous sequence. C1 shows the WT sequence, whereas C2 shows the heterozygous state. C3, only the insertion and the two nucleotide exchanges are shown after separating the WT sequence from the mutated sequence by gel electrophoresis. The red hyphens show the positions of the inserted nucleotides, whereas the red nucleotides represent the changed nucleotides. Black arrow, location of a nucleotide exchange; red arrow, position where the insertion starts. A: F1, c.166C>T, p.Gln56*; B: F3, c.140C>A, p.Ser47* p.Leu46Met; C: F2, c.133_134insGGATGTGCATTG, p.Lys45delinsArgMetCysIleGlu; c.135A>G, c.136C>A; D: F4, c.657T>A, p.Val216Glu; E: F5, c.676G>C, p.Gly226Arg.
Supplementary Figure 9 Sequence alignment of the proteins encoded by the zebrafish and human Stx1b orthologs.
An asterisk indicates a fully conserved residue. The colors provide information about the physicochemical properties of the individual amino acids. Different amino acids displaying the same color have similar physicochemical properties. A colon indicates strongly similar amino acid properties, and a period indicates weakly similar amino acid properties. Stx1b sequences were taken from the UniProt database (human, STX1B_HUMAN; zebrafish, Q1MTI1_DANRE). ClustalW2 2.1 was used to generate the sequence alignment.
Supplementary Figure 10 Synataxin-1B protein levels in zebrafish at 1 to 6 d.p.f. and images of larvae injected with a translation-blocking MO, stx1b-MO, at 5-d.p.f.
(a) Protein blot analysis of zebrafish Stx1b in wild-type embryos and larvae at 1 to 6 d.p.f. Stx1b was detected as early as 24 h.p.f., and its level gradually increased to reach a stable level at 4 to 5 d.p.f. (b) Dose-dependent knockdown of Stx1b in zebrafish embryos at 3 d.p.f. Injecting 3 and 6 ng of stx1b-MO resulted in a significant reduction in Stx1b protein. (c) Representative protein blots of stx1b-MO larvae from 1 to 6 d.p.f. At 1 d.p.f., complete knockdown was observed in the stx1b-MO larvae, and levels gradually decreased to reach a partial reduction in Stx1b protein at 5 and 6 d.p.f. GAPDH was used as a loading control for all experiments. (d) Quantification of syntaxin-1B. The most pronounced decrease in Stx1b expression was observed between 1 and 4 d.p.f. (13.6 ± 10.7%, 10.7 ± 6.1%, 13.2 ± 2.0% and 14.2 ± 2.4%, respectively). At 5 and 6 d.p.f., approximately 42.9 ± 18.1% and 67.2 ± 23.2% of the Stx1b protein could be detected. 5 d.p.f. was marked with a blue frame to emphasize the developmental stage at which all other experiments were performed. Quantification was performed on data obtained from 2 separate experiments normalized to the expression of Stx1b in ctrl MO–injected embryos. (e) Representative images of wild-type, ctrl-MO and stx1b-MO larvae at 5 d.p.f. Stx1b MO–injected larvae displayed aberrant morphologies such as absence of swim bladder (3, red arrow), pericardial edema (data not shown) or tail curvature (4, red arrow). Wild type (1) and ctrl-MO (2) lare developed normally.
Supplementary Figure 11 Confirmation of the stx1b-MO epileptiform phenotype using a second non-overlapping translation-blocking MO, stx1b-MO2.
(a) Stx1b protein levels in zebrafish knockdown embryos and larvae at 1 to 5 d.p.f. in a representative protein blot. A partial knockdown could be observed throughout all days analyzed in the stx1b-MO2–injected larvae. At 5 d.p.f., Stx1b reached a similar level as compared to stx1b-MO. GAPDH was used as a loading control. (b) Quantification of Stx1b protein blots. The decrease in Stx1b expression was observed during all 5 d (43.1 ± 3.4%, 19.9 ± 8.0%, 21.6 ± 3.7%, 38.0 ± 5.2%, 46.9 ± 12.0%, respectively), being the most pronounced at 5 d.p.f. Quantification was performed on data obtained from three separate experiments normalized to the expression of Stx1b in ctrl-MO–injected embryos. (c) Representative images of wild-type, ctrl-MO and stx1b-MO2 larvae at 5 d.p.f. stx1b-MO2–injected larvae displayed aberrant morphologies similar to those for stx1b-MO–injected larvae such as absence of swim bladder (3, red arrow) or pericardial edema (data not shown). No major abnormalities were observed in wild-type (1) and ctrl-MO (2) larvae. (d) Representative epileptiform activities of stx1b-MO2 larvae at 5 d.p.f. displaying polyspiking discharges (PD) (left) or high-frequency oscillations (HFO) (right). Top traces represent typical epileptiform patterns as seen in gap-free recordings. Bottom traces show high-resolution magnification of the epileptiform events mentioned above. Color maps represent the LFP frequency content of PD (left) and HFO (right) in the time-frequency domain. The color scale varies from blue (low magnitude) to red (high magnitude). The scales for the recording magnification and color maps are the same. LFP recordings were performed on larval optic tecta on stx1b-MO2 (n = 22) and ctrl-MO (n = 17) larvae at 5 d.p.f. Recurrent spontaneous epileptiform events occurred in 22 of 53 stx1b-MO2 knockdown larvae at a mean frequency of 7.6 ± 1.0 events/10 min (n = 22 stx1b-MO2 larvae analyzed; ****P < 0.0001), a mean duration of 260.3 ± 9.1 ms (n = 167 events analyzed; ****P < 0.0001) and a cumulative duration of seizures of 1,976 ± 276.3 ms/10 min (n = 22 stx1b-MO2 larvae analyzed; ****P < 0.0001), whereas controls only displayed baseline activity (data not shown).
Supplementary information
Combined Supplementary PDF
Supplementary Figures 1–11, Supplementary Tables 2–13 and Supplementary Note. (PDF 4577 kb)
Overview of the clinical details of the analyzed families.
Individuals are denoted according to the pedigrees in Figure 1. F female; M, male y, years; m, months; sf, seizure free; na, not applicable or available; G, German origin; D, Dutch origin; S, Swiss origin; GTCS, generalized tonic-clonic seizures; AbS, absence seizures; FS, febrile seizure; AtS, atonic seizures; TS, tonic seizures; CBZ, carbamazepine; CLB, clobazam; ESX, ethosuximide; LTG, lamotrigine; LCM, lacosamide; LEV, levetiracetame; PB, phenobarbital; PHT, phenytoin; PRM, primidone; VPA, valproate; STM, sultiam; OXC, oxcarbazepin; CBZ, carbamazepine; CLB, clobazam; LTG, lamotrigine; LEV, levetiracetame; RUF, rufinamide; LCM, lacosamide; ZSM, zonisamide; KD, ketogenic diet; VNS, vagus nerve stimulator. (XLSX 18 kb)
Ctrl-MO–injected larva at 5 d.p.f.
Touch response in ctrl-MO–injected larvae at 5 d.p.f. (AVI 30334 kb)
stx1b-MO–injected larv at 5 d.p.f.
No touch response in stx1b-MO–injected larva at 5 d.p.f. (AVI 35512 kb)
Ctrl-MO–injected larva at 5 d.p.f.
Normal behavior of ctrl-MO–injected larva at 5 d.p.f. (AVI 35512 kb)
stx1b-MO–injected larva 1 at 5 d.p.f.
Example of the stereotypical behavior of stx1b-MO–injected larvae at 5 d.p.f. (pectoral fin fluttering and increased orofacial movements). Such events occurred episodically, lasting seconds to, at maximum, less than a minute only in stx1b-knockdown zebrafish. (AVI 35512 kb)
stx1b-MO injected larva 2 at 5 d.p.f.
Example of the stereotypical behavior of stx1b-MO–injected larvae at 5 d.p.f. (myoclonus-like jerks). These events also occurred episodically, lasting seconds (here repeatedly, approximately every 10 s) only in stx1b-knockdown zebrafish. (AVI 35512 kb)
Rights and permissions
About this article
Cite this article
Schubert, J., Siekierska, A., Langlois, M. et al. Mutations in STX1B, encoding a presynaptic protein, cause fever-associated epilepsy syndromes. Nat Genet 46, 1327–1332 (2014). https://doi.org/10.1038/ng.3130
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3130
This article is cited by
-
Heterozygous and homozygous variants in STX1A cause a neurodevelopmental disorder with or without epilepsy
European Journal of Human Genetics (2023)
-
Syntabulin regulates neuronal excitation/inhibition balance and epileptic seizures by transporting syntaxin 1B
Cell Death Discovery (2023)
-
Studying ultra-rare variants in STX1A uncovers a novel neurodevelopmental disorder
European Journal of Human Genetics (2023)
-
Augmented impulsive behavior in febrile seizure-induced mice
Toxicological Research (2023)
-
The influence of genetics on epilepsy syndromes in infancy and childhood
Acta Epileptologica (2022)