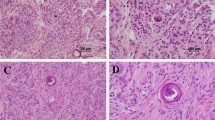
Abstract
Craniopharyngiomas are epithelial tumors that typically arise in the suprasellar region of the brain1. Patients experience substantial clinical sequelae from both extension of the tumors and therapeutic interventions that damage the optic chiasm, the pituitary stalk and the hypothalamic area2,3,4. Using whole-exome sequencing, we identified mutations in CTNNB1 (β-catenin) in nearly all adamantinomatous craniopharyngiomas examined (11/12, 92%) and recurrent mutations in BRAF (resulting in p.Val600Glu) in all papillary craniopharyngiomas (3/3, 100%). Targeted genotyping revealed BRAF p.Val600Glu in 95% of papillary craniopharyngiomas (36 of 39 tumors) and mutation of CTNNB1 in 96% of adamantinomatous craniopharyngiomas (51 of 53 tumors). The CTNNB1 and BRAF mutations were clonal in each tumor subtype, and we detected no other recurrent mutations or genomic aberrations in either subtype. Adamantinomatous and papillary craniopharyngiomas harbor mutations that are mutually exclusive and clonal. These findings have important implications for the diagnosis and treatment of these neoplasms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Louis, D.N., Ohgaki, H., Wiestler, O.D. & Cavenee, W.K. WHO Classification of Tumours of the Central Nervous System 238–240 (International Agency for Research on Cancer, 2007).
Crotty, T.B. et al. Papillary craniopharyngioma: a clinicopathological study of 48 cases. J. Neurosurg. 83, 206–214 (1995).
Duff, J. et al. Long-term outcomes for surgically resected craniopharyngiomas. Neurosurgery 46, 291–302, discussion 302–305 (2000).
Weiner, H.L. et al. Craniopharyngiomas: a clinicopathological analysis of factors predictive of recurrence and functional outcome. Neurosurgery 35, 1001–1010, discussion 1010–1011 (1994).
Dolecek, T.A., Propp, J.M., Stroup, N.E. & Kruchko, C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-oncol. 14 (suppl. 5), v1–v49 (2012).
Liubinas, S.V., Munshey, A.S. & Kaye, A.H. Management of recurrent craniopharyngioma. J. Clin. Neurosci. 18, 451–457 (2011).
Manley, P.E. et al. Sleep dysfunction in long term survivors of craniopharyngioma. J. Neurooncol. 108, 543–549 (2012).
Barkhoudarian, G. & Laws, E.R. Craniopharyngioma: history. Pituitary 16, 1–8 (2013).
Cushing, H. Intracranial tumors: notes upon a series of two thousand cases with surgical mortality percentages pertaining thereto. JAMA 100, 284 (1932).
Buslei, R. et al. Common mutations of β-catenin in adamantinomatous craniopharyngiomas but not in other tumours originating from the sellar region. Acta Neuropathol. 109, 589–597 (2005).
Kato, K. et al. Possible linkage between specific histological structures and aberrant reactivation of the Wnt pathway in adamantinomatous craniopharyngioma. J. Pathol. 203, 814–821 (2004).
Sekine, S. et al. Craniopharyngiomas of adamantinomatous type harbor β-catenin gene mutations. Am. J. Pathol. 161, 1997–2001 (2002).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Lawrence, M.S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Brastianos, P.K. et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat. Genet. 45, 285–289 (2013).
Futreal, P.A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004).
Stanley, F.K., Moore, S. & Goodarzi, A.A. CHD chromatin remodelling enzymes and the DNA damage response. Mutat. Res. 750, 31–44 (2013).
Laczmanska, I. & Sasiadek, M.M. Tyrosine phosphatases as a superfamily of tumor suppressors in colorectal cancer. Acta Biochim. Pol. 58, 467–470 (2011).
Jones, D.T. et al. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene 28, 2119–2123 (2009).
Tian, Y. et al. Detection of KIAA1549-BRAF fusion transcripts in formalin-fixed paraffin-embedded pediatric low-grade gliomas. J. Mol. Diagn. 13, 669–677 (2011).
Capper, D. et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 122, 11–19 (2011).
Anastas, J.N. & Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 13, 11–26 (2013).
Basu, A. et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151–1161 (2013).
Chapman, P.B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011).
Flaherty, K.T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010).
Ribas, A. & Flaherty, K.T. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat. Rev. Clin. Oncol. 8, 426–433 (2011).
Sosman, J.A. et al. Survival in BRAF V600–mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 366, 707–714 (2012).
Chamberlain, M.C. Salvage therapy with BRAF inhibitors for recurrent pleomorphic xanthoastrocytoma: a retrospective case series. J. Neurooncol. 114, 237–240 (2013).
Rush, S., Foreman, N. & Liu, A. Brainstem ganglioglioma successfully treated with vemurafenib. J. Clin. Oncol. 31, e159–e160 (2013).
Ascierto, P.A. et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J. Clin. Oncol. 31, 3205–3211 (2013).
Sievert, A.J. et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc. Natl. Acad. Sci. USA 110, 5957–5962 (2013).
Dietrich, S. et al. BRAF inhibition in refractory hairy-cell leukemia. N. Engl. J. Med. 366, 2038–2040 (2012).
Dias-Santagata, D. et al. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: diagnostic and therapeutic implications. PLoS ONE 6, e17948 (2011).
Schindler, G. et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 121, 397–405 (2011).
MacConaill, L.E. et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS ONE 4, e7887 (2009).
Demichelis, F. et al. SNP panel identification assay (SPIA): a genetic-based assay for the identification of cell lines. Nucleic Acids Res. 36, 2446–2456 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Chapman, M.A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011).
Berger, M.F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011).
Lohr, J.G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 109, 3879–3884 (2012).
Stransky, N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011).
Carter, S.L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Rickert, C.H. & Paulus, W. Lack of chromosomal imbalances in adamantinomatous and papillary craniopharyngiomas. J. Neurol. Neurosurg. Psychiatry 74, 260–261 (2003).
Yoshimoto, M. et al. Comparative genomic hybridization analysis of pediatric adamantinomatous craniopharyngiomas and a review of the literature. J. Neurosurg. 101, 85–90 (2004).
Landau, D.A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013).
Thomas, R.K. et al. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 39, 347–351 (2007).
Corcoran, R.B. et al. BRAF gene amplification can promote acquired resistance to MEKinhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal. 3, ra84 (2010).
Dias-Santagata, D. et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol. Med. 2, 146–158 (2010).
Acknowledgements
We thank M. Ducar for his assistance with genomic analyses; S. Chauvin for project management; L. Brown and H. Malkin for assisting with sample collection; T. Woo, B. Rich, R. Machaidze and D. Feldman for technical assistance; N. Stransky for the design of Figure 2a; and H. Taylor-Weiner and C.H. Brastianos for critical review of the manuscript. This work was supported by the Jared Branfman Sunflowers for Life Fund for Pediatric Brain and Spinal Cancer Research, the Pediatric Low-Grade Astrocytoma (PLGA) Program (S.S., W.C.H. and Charles D. Stiles), Pedals for Pediatrics and the Clark Family (P.E.M. and M.W.K.), the Stahl Family Charitable Foundation (P.E.M.), the Stop & Shop Pediatric Brain Tumor Program (P.E.M. and M.W.K.), the Pediatric Brain Tumor Clinical and Research Fund (P.E.M. and M.W.K.), the Children's Brain Tumor Foundation and the V Foundation (S.S.). S.S. is supported by grant K08 NS064168, and P.K.B. is supported by grant K12 CA090354-11, the Brain Science Foundation, Susan G. Komen for the Cure, the Terri Brodeur Breast Cancer Foundation, the Conquer Cancer Foundation and the American Brain Tumor Association.
Author information
Authors and Affiliations
Contributions
P.K.B., P.E.M., M.W.K., G.G. and S.S. designed the study. P.K.B., A.T.-W., C.S., G.G. and S.S. wrote the manuscript. A.T.-W., P.K.B., C.S., A.R.T. and G.G. performed computational analyses. P.V.H. supervised the sequencing. S.S. and D.N.L. reviewed the histopathology, and S.S., D.N.L. and M.P.H. coordinated and reviewed the immunohistochemistry. K.L.L. managed the tissue repository. R.T.J., L.A.B., A.S., N.S., L.S. and M.L.C. coordinated sample acquisition, processed samples and coordinated and performed exome and targeted sequencing. P.K.B., W.C.H., D.D.-S., D.N.L., A.C.R., M.W.K., G.G. and S.S. supervised the study. H.G.W.L., E.R.L., I.F.D., R.M.S., P.B.S., J.Y.K.L., J.N.P., N.D.A., H.T., M.M.-L., M.S., F.J.R., P.E.M., A.C.R., D.D.-S., D.N.L. and S.S. identified and provided materials for sequencing and validation, as well as clinical information. M.S.L. provided the code and the data to generate Figure 1. All authors discussed the results and implications and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Tables 1–4
Supplementary Tables 1–4 (XLSX 307 kb)
Rights and permissions
About this article
Cite this article
Brastianos, P., Taylor-Weiner, A., Manley, P. et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat Genet 46, 161–165 (2014). https://doi.org/10.1038/ng.2868
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2868
This article is cited by
-
Integrating Systemic Therapies into the Multimodality Therapy of Patients with Craniopharyngioma
Current Treatment Options in Oncology (2024)
-
Reproducible and clinically translatable deep neural networks for cervical screening
Scientific Reports (2023)
-
Shifting Strategies in the Treatment of Pediatric Craniopharyngioma
Current Oncology Reports (2023)
-
Spinal ectopic recurrence of craniopharyngioma in a pediatric patient
Child's Nervous System (2023)
-
Dabrafenib as a diagnostic and therapeutic strategy for the non-surgical management of papillary craniopharyngioma
Pituitary (2023)