Abstract
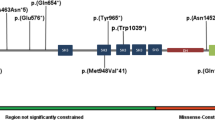
Prader-Willi syndrome (PWS) is caused by the absence of paternally expressed, maternally silenced genes at 15q11-q13. We report four individuals with truncating mutations on the paternal allele of MAGEL2, a gene within the PWS domain. The first subject was ascertained by whole-genome sequencing analysis for PWS features. Three additional subjects were identified by reviewing the results of exome sequencing of 1,248 cases in a clinical laboratory. All four subjects had autism spectrum disorder (ASD), intellectual disability and a varying degree of clinical and behavioral features of PWS. These findings suggest that MAGEL2 is a new gene causing complex ASD and that MAGEL2 loss of function can contribute to several aspects of the PWS phenotype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Holm, V.A. et al. Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics 91, 398–402 (1993).
Cassidy, S.B., Schwartz, S., Miller, J.L. & Driscoll, D.J. Prader-Willi syndrome. Genet. Med. 14, 10–26 (2012).
Sahoo, T. et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 40, 719–721 (2008).
de Smith, A.J. et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 18, 3257–3265 (2009).
Duker, A.L. et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur. J. Hum. Genet. 18, 1196–1201 (2010).
Kanber, D. et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur. J. Hum. Genet. 17, 582–590 (2009).
Resnick, J.L., Nicholls, R.D. & Wevrick, R. Recommendations for the investigation of animal models of Prader-Willi syndrome. Mamm. Genome 24, 165–178 (2013).
Tennese, A.A. & Wevrick, R. Impaired hypothalamic regulation of endocrine function and delayed counterregulatory response to hypoglycemia in Magel2-null mice. Endocrinology 152, 967–978 (2011).
Mercer, R.E. & Wevrick, R. Loss of Magel2, a candidate gene for features of Prader-Willi syndrome, impairs reproductive function in mice. PLoS ONE 4, e4291 (2009).
Bischof, J.M., Stewart, C.L. & Wevrick, R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum. Mol. Genet. 16, 2713–2719 (2007).
Drmanac, R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010).
Doyle, J.M., Gao, J., Wang, J., Yang, M. & Potts, P.R. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell 39, 963–974 (2010).
Peters, B.A. et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature 487, 190–195 (2012).
Leung, K.N., Vallero, R.O., DuBose, A.J., Resnick, J.L. & LaSalle, J.M. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum. Mol. Genet. 18, 4227–4238 (2009).
Descheemaeker, M.J., Govers, V., Vermeulen, P. & Fryns, J.P. Pervasive developmental disorders in Prader-Willi syndrome: the Leuven experience in 59 subjects and controls. Am. J. Med. Genet. A. 140, 1136–1142 (2006).
O'Roak, B.J. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43, 585–589 (2011).
Sanders, S.J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012).
O'Roak, B.J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012).
Neale, B.M. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012).
Iossifov, I. et al. De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299 (2012).
O'Roak, B.J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Michaelson, J.J. et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 (2012).
Yu, T.W. et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron 77, 259–273 (2013).
Bervini, S. & Herzog, H. Mouse models of Prader-Willi Syndrome: a systematic review. Front. Neuroendocrinol. 34, 107–119 (2013).
Huang, H.S. et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481, 185–189 (2012).
Berteaux, N. et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol. Cell. Biol. 28, 6731–6745 (2008).
Boone, P.M. et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum. Mutat. 31, 1326–1342 (2010).
Acknowledgements
We are indebted to the patients and their families for their willingness to participate in our research study. We thank P. Zimmerman and E. Austin for clinical assistance. C.P.S. is generously supported by the Joan and Stanford Alexander family. C.P.S. is a recipient of a Clinical Scientist Development Award from the Doris Duke Charitable Foundation. M.L.G.-G. and C.T.C. are generously supported by the Cullen Foundation for Higher Education and the Houston Foundation. A.L.B. is supported by US National Institutes of Health grant HD037283.
Author information
Authors and Affiliations
Contributions
M.L.G.-G. and M.A.M. performed whole-genome sequencing and phase determination on subject 1. M.L.G.-G., M.A.M., B.A.P., R.D. and C.T.C. designed and analyzed the experiments for subject 1. F.X. and Y.Y. performed whole-exome sequencing and phase determination on subjects 2–4. C.P.S., F.X., Y.Y., B.Z., A.L.B. and Y.Y. designed and analyzed the experiments for subjects 2–4. L.P. and K.W.G. contributed subjects and provided detailed physical examinations. C.P.S. conceived the overall study, coordinated enrollment, supervised the experiments, wrote the manuscript and generated the figures and tables. All authors participated in the discussion and interpretation of data and results, and all participated in editing and revising the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.P.S., A.L.B., C.T.C. and Y.Y. are faculty members of the Department of Molecular and Human Genetics at the Baylor College of Medicine, which derives revenue from whole-exome sequencing analysis offered in the Medical Genetics Laboratory. B.A.P., M.A.M. and R.D. are employees of Complete Genomics, a company that derives revenue from whole-genome sequencing analysis. Complete Genomics has filed several patents on sequencing technology. The remaining authors declare no conflict of interest.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1 and 2, and Supplementary Tables 1 and 2 (PDF 3503 kb)
Rights and permissions
About this article
Cite this article
Schaaf, C., Gonzalez-Garay, M., Xia, F. et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet 45, 1405–1408 (2013). https://doi.org/10.1038/ng.2776
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2776
This article is cited by
-
Direct haplotype-resolved 5-base HiFi sequencing for genome-wide profiling of hypermethylation outliers in a rare disease cohort
Nature Communications (2023)
-
Oxytocin-based therapies for treatment of Prader-Willi and Schaaf-Yang syndromes: evidence, disappointments, and future research strategies
Translational Psychiatry (2022)
-
A nationwide survey of Schaaf-Yang syndrome in Japan
Journal of Human Genetics (2022)
-
Early life oxytocin treatment improves thermo-sensory reactivity and maternal behavior in neonates lacking the autism-associated gene Magel2
Neuropsychopharmacology (2022)
-
The contribution of imprinted genes to neurodevelopmental and neuropsychiatric disorders
Translational Psychiatry (2022)