Abstract
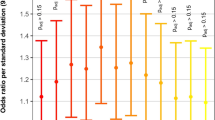
Congenital heart malformation (CHM) is the most common form of congenital human birth anomaly and is the leading cause of infant mortality. Although some causative genes have been identified, little progress has been made in identifying genes in which low-penetrance susceptibility variants occur in the majority of sporadic CHM cases. To identify common genetic variants associated with sporadic non-syndromic CHM in Han Chinese populations, we performed a multistage genome-wide association study (GWAS) in a total of 4,225 CHM cases and 5,112 non-CHM controls. The GWAS stage included 945 cases and 1,246 controls and was followed by 2-stage validation with 2,160 cases and 3,866 controls. The combined analyses identified significant associations (P < 5.0 × 10−8) at 1p12 (rs2474937 near TBX15; odds ratio (OR) = 1.40; P = 8.44 × 10−10) and 4q31.1 (rs1531070 in MAML3; OR = 1.40; P = 4.99 × 10−12). These results extend current knowledge of genetic contributions to CHM in Han Chinese populations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hoffman, J.I. & Kaplan, S. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 (2002).
Bruneau, B.G. The developmental genetics of congenital heart disease. Nature 451, 943–948 (2008).
Krebs, L.T. et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343–1352 (2000).
Wang, J., Greene, S.B. & Martin, J.F. BMP signaling in congenital heart disease: new developments and future directions. Birth Defects Res. A Clin. Mol. Teratol. 91, 441–448 (2011).
Anderson, L.M. & Gibbons, G.H. Notch: a mastermind of vascular morphogenesis. J. Clin. Invest. 117, 299–302 (2007).
Olson, E.N. Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927 (2006).
Srivastava, D. Making or breaking the heart: from lineage determination to morphogenesis. Cell 126, 1037–1048 (2006).
Pierpont, M.E. et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115, 3015–3038 (2007).
Weismann, C.G. & Gelb, B.D. The genetics of congenital heart disease: a review of recent developments. Curr. Opin. Cardiol. 22, 200–206 (2007).
Øyen, N. et al. Recurrence of congenital heart defects in families. Circulation 120, 295–301 (2009).
Burn, J. et al. Recurrence risks in offspring of adults with major heart defects: results from first cohort of British collaborative study. Lancet 351, 311–316 (1998).
Wessels, M.W. & Willems, P.J. Genetic factors in non-syndromic congenital heart malformations. Clin. Genet. 78, 103–123 (2010).
Ware, S.M. & Jefferies, J.L. New genetic insights into congenital heart disease. J. Clin. Exp. Cardiolog. S8, pii: 003 (2012).
Roessler, E. et al. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am. J. Hum. Genet. 83, 18–29 (2008).
Van Driel, L.M. et al. Eight-fold increased risk for congenital heart defects in children carrying the nicotinamide N-methyltransferase polymorphism and exposed to medicines and low nicotinamide. Eur. Heart J. 29, 1424–1431 (2008).
van Beynum, I.M. et al. Common 894G>T single nucleotide polymorphism in the gene coding for endothelial nitric oxide synthase (eNOS) and risk of congenital heart defects. Clin. Chem. Lab. Med. 46, 1369–1375 (2008).
Xu, J. et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum. Mutat. 30, 1231–1236 (2009).
Zhao, J. et al. The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature. BMC Med. Genet. 11, 96 (2010).
Kim, J.J. et al. Identification of 15 loci influencing height in a Korean population. J. Hum. Genet. 55, 27–31 (2010).
Heid, I.M. et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42, 949–960 (2010).
Li, Q.Y. et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet. 15, 21–29 (1997).
Basson, C.T. et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 15, 30–35 (1997).
Borozdin, W. et al. Expanding the spectrum of TBX5 mutations in Holt-Oram syndrome: detection of two intragenic deletions by quantitative real time PCR, and report of eight novel point mutations. Hum. Mutat. 27, 975–976 (2006).
Plageman, T.F. Jr. & Yutzey, K.E. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J. Biol. Chem. 279, 19026–19034 (2004).
Jerome, L.A. & Papaioannou, V.E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 27, 286–291 (2001).
Lindsay, E.A. et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410, 97–101 (2001).
Merscher, S. et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104, 619–629 (2001).
Stennard, F.A. & Harvey, R.P. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development 132, 4897–4910 (2005).
Kirk, E.P. et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am. J. Hum. Genet. 81, 280–291 (2007).
Qian, L. et al. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc. Natl. Acad. Sci. USA 105, 19833–19838 (2008).
Posch, M.G. et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J. Med. Genet. 47, 230–235 (2010).
Hadjantonakis, A.K., Pisano, E. & Papaioannou, V.E. Tbx6 regulates left/right patterning in mouse embryos through effects on nodal cilia and perinodal signaling. PLoS ONE 3, e2511 (2008).
Christoffels, V.M. et al. Formation of the venous pole of the heart from an Nkx2–5-negative precursor population requires Tbx18. Circ. Res. 98, 1555–1563 (2006).
Habets, P.E. et al. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 16, 1234–1246 (2002).
Singh, M.K. et al. The T-box transcription factor Tbx15 is required for skeletal development. Mech. Dev. 122, 131–144 (2005).
Lausch, E. et al. TBX15 mutations cause craniofacial dysmorphism, hypoplasia of scapula and pelvis, and short stature in Cousin syndrome. Am. J. Hum. Genet. 83, 649–655 (2008).
Gesta, S. et al. Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc. Natl. Acad. Sci. USA 108, 2771–2776 (2011).
Meins, M., Henderson, D.J., Bhattacharya, S.S. & Sowden, J.C. Characterization of the human TBX20 gene, a new member of the T-Box gene family closely related to the Drosophila H15 gene. Genomics 67, 317–332 (2000).
Naiche, L.A., Harrelson, Z., Kelly, R.G. & Papaioannou, V.E. T-box genes in vertebrate development. Annu. Rev. Genet. 39, 219–239 (2005).
Wehn, A.K. & Chapman, D.L. Tbx18 and Tbx15 null-like phenotypes in mouse embryos expressing Tbx6 in somitic and lateral plate mesoderm. Dev. Biol. 347, 404–413 (2010).
Artavanis-Tsakonas, S., Matsuno, K. & Fortini, M.E. Notch signaling. Science 268, 225–232 (1995).
Oyama, T. et al. Mastermind-1 is required for Notch signal-dependent steps in lymphocyte development in vivo. Proc. Natl. Acad. Sci. USA 104, 9764–9769 (2007).
Oyama, T. et al. Mastermind-like 1 (MamL1) and mastermind-like 3 (MamL3) are essential for Notch signaling in vivo. Development 138, 5235–5246 (2011).
Kraft, P. Curses—winner's and otherwise—in genetic epidemiology. Epidemiology 19, 649–651, discussion 657–658 (2008).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Acknowledgements
The authors wish to thank all the study participants, research staff and students who participated in this work and Q. Wei (University of Texas MD Anderson Cancer Center) for his scientific editing of the manuscript. This work was partly funded by the National Key Basic Research Program Grant (2011CB944304 and 2012CB944902), the 863 Program (2012AA02A515), a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine), the Natural Science Foundation of China (81000076, 81130022 and 81121001) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT1025).
Author information
Authors and Affiliations
Contributions
H.S., J.S., Y.C., Z.Z. and Z.H. directed the study, obtained financial support and were responsible for study design, the interpretation of results and manuscript writing. Y.S. directed the GWAS. J.D. and Y.L. were responsible for statistical analyses. Y.L., S.P., M.D., B.Q., Y.W. and J.W. were responsible for sample processing and managed the genotyping data. X.M., J. Xu, S. Yang, Z.X., Xiaowei Wang, X.G., Y.X., H.M., G.J., S. Yu, J.L. and Xinru Wang were responsible for subject recruitment and sample preparation for the Nanjing samples. B.Z. and J. Xing were responsible for subject recruitment and sample preparation for the Xi'an samples. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–9 (PDF 1571 kb)
Rights and permissions
About this article
Cite this article
Hu, Z., Shi, Y., Mo, X. et al. A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations. Nat Genet 45, 818–821 (2013). https://doi.org/10.1038/ng.2636
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2636
This article is cited by
-
Association between MTHFR C677T variant and risk for congenital heart defects in Egyptian children: a case–control study including meta-analysis based on 147 cases and 143 controls
Egyptian Journal of Medical Human Genetics (2023)
-
Loss of GLTSCR1 causes congenital heart defects by regulating NPPA transcription
Angiogenesis (2023)
-
Functional screening of congenital heart disease risk loci identifies 5 genes essential for heart development in zebrafish
Cellular and Molecular Life Sciences (2023)
-
Identification and functional analysis of variants of MYH6 gene promoter in isolated ventricular septal defects
BMC Medical Genomics (2022)
-
Genomic frontiers in congenital heart disease
Nature Reviews Cardiology (2022)