Abstract
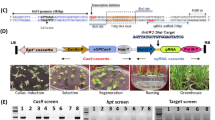
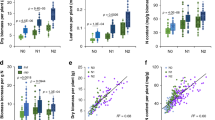
Increases in the yield of rice, a staple crop for more than half of the global population, are imperative to support rapid population growth. Grain weight is a major determining factor of yield. Here, we report the cloning and functional analysis of THOUSAND-GRAIN WEIGHT 6 (TGW6), a gene from the Indian landrace rice Kasalath. TGW6 encodes a novel protein with indole-3-acetic acid (IAA)-glucose hydrolase activity. In sink organs, the Nipponbare tgw6 allele affects the timing of the transition from the syncytial to the cellular phase by controlling IAA supply and limiting cell number and grain length. Most notably, loss of function of the Kasalath allele enhances grain weight through pleiotropic effects on source organs and leads to significant yield increases. Our findings suggest that TGW6 may be useful for further improvements in yield characteristics in most cultivars.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sweeney, M. & McCouch, S. The complex history of the domestication of rice. Ann. Bot. (Lond.) 100, 951–957 (2007).
Harlan, J. Crops and Man (American Society of Agronomy, Crop Science Society of America, Madison, Wisconsin, 1992).
Fan, C. et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171 (2006).
Takano-Kai, N. et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182, 1323–1334 (2009).
Shomura, A. et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028 (2008).
Wang, S. et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954 (2012).
Nagata, K., Yoshinaga, S., Takanashi, J. & Terao, T. Effects of dry matter production, translocation of nonstructural carbohydrates and nitrogen application on grain filling in rice cultivar Takanari, a cultivar bearing a large number of spikelets. Plant Prod. Sci. 4, 173–183 (2001).
Yang, J. et al. Grain and dry matter yields and partitioning of assimilates in japonica/indica hybrid rice. Crop Sci. 42, 766–772 (2002).
Peng, S., Khush, G.S., Virk, P., Tang, Q. & Zoh, Y. Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 108, 32–38 (2008).
Dingkuhn, M., Penning de Vries, F.M.T., de Datta, S.K. & van Laar, H.H. Concepts for a New Plant Type for Direct Seeded Flooded Tropical Rice (International Rice Research Institute, Los Baños, Philippines, 1991).
Ishimaru, K., Kashiwagi, T., Hirotsu, N. & Madoka, Y. Identification and physiological analyses of a locus for rice yield potential across the genetic background. J. Exp. Bot. 56, 2745–2753 (2005).
Ishimaru, K., Kosone, M., Sasaki, H. & Kashiwagi, T. Leaf contents differ depending on the position in a rice leaf sheath during sink-source transition. Plant Physiol. Biochem. 42, 855–860 (2004).
Ishimaru, K. Identification of a locus increasing rice yield and physiological analysis of its function. Plant Physiol. 133, 1083–1090 (2003).
Hoshikawa, K. The Growing Rice Plant: an Anatomical Monograph (Nosan Gyoson Bunka Kyokai, Tokyo, 1989).
Hoshikawa, K. Studies on the development in rice. Process of endosperm tissue formation. Jpn. J. Crop. Sci. 36, 151–161 (1967).
Brown, R.C., Lemmon, B.E. & Olsen, O.-D. Development of the endosperm in rice (Oryza sativa L.): cellularization. J. Plant Res. 109, 301–313 (1996).
Mizutani, M., Naganuma, T., Tsutsumi, K. & Saitoh, Y. The syncytium-specific expression of the Orysa;KRP3 CDK inhibitor: implication of its involvement in the cell cycle control in the rice (Oryza sativa L.) syncytial endosperm. J. Exp. Bot. 61, 791–798 (2010).
Jawad, Z. & Paoli, M. Novel sequences propel familiar folds. Structure 10, 447–454 (2002).
Marchler-Bauer, A. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 (2011).
Scharff, E.I., Koepke, J., Fritzsch, G., Lucke, C. & Ruterjans, H. Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris. Structure 9, 493–502 (2001).
Lur, H.S. & Setter, T.L. Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol. 103, 273–280 (1993).
Ludwig-Müller, J. Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773 (2011).
Jakubowska, A. & Kowalczyk, S. A specific enzyme hydrolyzing 6-O(4-O)-indole-3-ylacetyl-β-D-glucose in immature kernels of Zea mays. J. Plant Physiol. 162, 207–213 (2005).
Song, X.-J. et al. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630 (2007).
Li, Y. et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269 (2011).
Wang, E. et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40, 1370–1374 (2008).
Kurata, N. & Yamazaki, Y. Oryzabase. an integrated biological and genome information database for rice. Plant Physiol. 140, 12–17 (2006).
Kojima, Y., Ebana, K., Fukuoka, S., Nagamine, T. & Kawase, M. Development of an RFLP-based rice diversity research set of germplasm. Breed. Sci. 55, 431–440 (2005).
Tanksley, S.D. & McCouch, S.R. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066 (1997).
Yamakawa, H., Hirose, T., Kuroda, M. & Yamaguchi, T. Comprehensive expression profiling of rice grain filling–related genes under high temperature using DNA microarray. Plant Physiol. 144, 258–277 (2007).
Peng, S. et al. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 101, 9971–9975 (2004).
Miki, D. & Shimamoto, K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495 (2004).
Toki, S. et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976 (2006).
Ma, X., Panjikar, S., Koepke, J., Loris, E. & Stöckigt, J. The sturucture of Rauvolfia serpentina strictosidine synthase is novel six-bladed β-propeller fold in plant proteins. Plant Cell 18, 907–920 (2006).
Stöckigt, J., Barleben, L., Panjikar, S. & Loris, E.A. 3D-structure and function of strictosidine synthase—the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol. Biochem. 46, 340–355 (2008).
Shi, J., Blundell, T.L. & Mizuguchi, K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257 (2001).
Dolan, M.A., Keil, M. & Baker, D.S. Comparison of composer and ORCHESTRAR. Proteins 72, 1243–1258 (2008).
Clark, M., Cramer, R.D. & van den Opdenbosch, N. III. Validation of the general purpose tripose 5.2 force field. J. Comput. Chem. 10, 982–1012 (1989).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
Tsunoda, Y. et al. Improving expression and solubility of rice proteins produced as fusion proteins in Escherichia coli. Protein Expr. Purif. 42, 268–277 (2005).
Ishimaru, K., Hirotsu, N., Madoka, Y. & Kashiwagi, T. Quantitative trait loci for sucrose, starch, and hexose accumulation before heading in rice. Plant Physiol. Biochem. 45, 799–804 (2007).
Matsuda, F., Miyazawa, H., Wakasa, K. & Miyagawa, H. Quantification of indole-3-acetic acid and amino acid conjugates in rice by liquid chromatography–electrospray ionization–tandem mass spectrometry. Biosci. Biotechnol. Biochem. 69, 778–783 (2005).
Hirose, T. et al. Expression profiling of genes related to starch synthesis in rice leaf sheaths during the heading period. Physiol. Plant. 128, 425–435 (2006).
Komatsuda, T. et al. Six-rowed barley originated from a mutation in a homeodomain–leucine zipper I–class homeobox gene. Proc. Natl. Acad. Sci. USA 104, 1424–1429 (2007).
Clement, M., Posada, D. & Crandall, K. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 (2000).
Acknowledgements
We thank T. Meshi for helpful and important suggestions, Y. Shimoda and A. Miyamoto for help with experiments, T. Yanai and S. Yoshimizu for rice growth, K. Shimamoto (Nara Institute of Science and Technology) for providing the pANDA vector, K. Toshimitsu (Japan International Cooperation Agency) for providing seeds of NERICA and T. Kobayakawa for helpful discussions. The wild rice accessions used in this study were distributed from the National Institute of Genetics supported by the National Bioresource Project, Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was supported by grants from the Bio Cosmos Program and Genomics for Agricultural Innovation (QTL-4007), Ministry of Agriculture, Forestry and Fisheries of Japan.
Author information
Authors and Affiliations
Contributions
K.I. conceived the research project, designed experiments and wrote the manuscript with help from N. Hirotsu, Y.M. and E.K. K.I., N. Hirotsu and Y.M. carried out field phenotyping, genetics, gene cloning and functional and molecular evolution experiments and analyzed the data. N. Hara helped with in situ hybridization, and H.O. assisted in transformation. N.M. helped with phenotyping, genotyping and gene cloning. T.K. and K.U. helped with field work, and B.S., A.O. and H.M. quantified IAA content. E.K. performed structure analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–21 and Supplementary Tables 1–5 (PDF 2839 kb)
Rights and permissions
About this article
Cite this article
Ishimaru, K., Hirotsu, N., Madoka, Y. et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45, 707–711 (2013). https://doi.org/10.1038/ng.2612
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2612
This article is cited by
-
Development of introgression lines and mapping of qGW2, a novel QTL that confers grain width, in rice (Oryza sativa L.)
Molecular Breeding (2024)
-
Genetic and functional mechanisms of yield-related genes in rice
Acta Physiologiae Plantarum (2024)
-
Genetic diversity of grain yield traits and identification of a grain weight gene SiTGW6 in foxtail millet
Theoretical and Applied Genetics (2024)
-
Identification of qGL4.1 and qGL4.2, two closely linked QTL controlling grain length in rice
Molecular Breeding (2024)
-
Can the Wild Perennial, Rhizomatous Rice Species Oryza longistaminata be a Candidate for De Novo Domestication?
Rice (2023)