Abstract
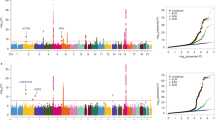
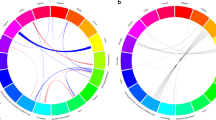
We conducted a genome-wide association study (GWAS) and a genome-wide gene-environment interaction analysis of esophageal squamous-cell carcinoma (ESCC) in 2,031 affected individuals (cases) and 2,044 controls with independent validation in 8,092 cases and 8,620 controls. We identified six new ESCC susceptibility loci, of which four, at chromosomes 4q23, 16q12.1, 22q12 and 3q27 had a significant marginal effect (P = 1.78 × 10−39 to P = 2.49 × 10−11) and two of which, at 2q22 and 13q33, had a significant association only in the gene–alcohol drinking interaction (gene-environment interaction P (PG × E) = 4.39 × 10−11 and PG × E = 4.80 × 10−8, respectively). Variants at the 4q23 locus, which includes the ADH cluster, each had a significant interaction with alcohol drinking in their association with ESCC risk (PG × E = 2.54 × 10−7 to PG × E = 3.23 × 10−2). We confirmed the known association of the ALDH2 locus on 12q24 to ESCC, and a joint analysis showed that drinkers with both of the ADH1B and ALDH2 risk alleles had a fourfold increased risk for ESCC compared to drinkers without these risk alleles. Our results underscore the direct genetic contribution to ESCC risk, as well as the genetic contribution to ESCC through interaction with alcohol consumption.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
27 August 2014
In the version of this article initially published, in Table 1 and its associated text, there was a calculation error in which the relative sizes of the case and control populations were set to be equal; because the size of the case population (15,767) was nearly double that of the control population (8,329), this resulted in erroneously inflated penetrance estimates. A simple definition of penetrance is used that is often applied in medical genetics—namely, the proportion of observed mutation carriers that are affected—to provide a metric that would be useful to clinical geneticists in a setting in which disease is heavily enriched, for example, in diagnosing children with developmental delay. That formulation is biased upwards with respect to population-level penetrance. Thus, in this corrigendum, an estimate more appropriate for population-level inference is provided assuming a general disease prevalence of 5.3% (Am. J. Hum. Genet. 42, 677–693, 1988) along with the more familiar odds ratio (OR) estimate. Importantly, all of these measures of penetrance are intrinsically limited by sampling error and imprecision in defining disease prevalence. We note that the mutation carrier counts, P values and other results in the original version of Table 1 are correct, and the key results and conclusions of the paper are unaffected. The error has been corrected in the HTML and PDF versions of the article.
References
Sun, T. et al. Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J. Natl. Cancer Inst. 96, 1030–1036 (2004).
Zhang, X. et al. Identification of functional genetic variants in cyclooxygenase-2 and their association with risk of esophageal cancer. Gastroenterology 129, 565–576 (2005).
Sun, T. et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat. Genet. 39, 605–613 (2007).
Lewis, S.J. & Smith, G.D. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol. Biomarkers Prev. 14, 1967–1971 (2005).
Hiyama, T., Yoshihara, M., Tanaka, S. & Chayama, K. Genetic polymorphisms and esophageal cancer risk. Int. J. Cancer 121, 1643–1658 (2007).
Akbari, M.R. et al. Candidate gene association study of esophageal squamous cell carcinoma in a high-risk region in Iran. Cancer Res. 69, 7994–8000 (2009).
Hashibe, M. et al. Multiple ADH genes are associated with upper aerodigestive tract cancers. Nat. Genet. 40, 707–709 (2008).
McKay, J.D. et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 7, e1001333 (2011).
Cui, R. et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 137, 1768–1775 (2009).
Wang, L.D. et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat. Genet. 42, 759–763 (2010).
Abnet, C.C. et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet. 42, 764–767 (2010).
Wu, C. et al. Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat. Genet. 43, 679–684 (2011).
Panagiotou, O.A. & Ioannidis, J.P. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int. J. Epidemiol. 41, 273–286 (2012).
Park, J.H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Islami, F. et al. Alcohol drinking and esophageal squamous cell carcinoma with focus on light-drinkers and never-smokers: a systematic review and meta-analysis. Int. J. Cancer 129, 2473–2484 (2011).
Hunter, D.J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 6, 287–298 (2005).
Gao, Y.T. et al. Risk factors for esophageal cancer in Shanghai, China. I. Role of cigarette smoking and alcohol drinking. Int. J. Cancer 58, 192–196 (1994).
Kim, R., Emi, M., Tanabe, K. & Murakami, S. Role of the unfolded protein response in cell death. Apoptosis 11, 5–13 (2006).
Koong, A.C., Chauhan, V. & Romero-Ramirez, L. Targeting XBP-1 as a novel anti-cancer strategy. Cancer Biol. Ther. 5, 756–759 (2006).
Romero-Ramirez, L. et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 64, 5943–5947 (2004).
Shuda, M. et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J. Hepatol. 38, 605–614 (2003).
Antoni, L., Sodha, N., Collins, I. & Garrett, M.D. CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat. Rev. Cancer 7, 925–936 (2007).
Dall′Olio, F., Chiricolo, M. & Lau, J.T. Differential expression of the hepatic transcript of β-galactoside α-2,6-sialyltransferase in human colon cancer cell lines. Int. J. Cancer 81, 243–247 (1999).
Wang, P.H. et al. Enhanced expression of α 2,6-sialyltransferase ST6Gal I in cervical squamous cell carcinoma. Gynecol. Oncol. 89, 395–401 (2003).
Recchi, M.A. et al. Multiplex reverse transcription polymerase chain reaction assessment of sialyltransferase expression in human breast cancer. Cancer Res. 58, 4066–4070 (1998).
Pousset, D., Piller, V., Bureaud, N., Monsigny, M. & Piller, F. Increased α 2,6 sialylation of N-glycans in a transgenic mouse model of hepatocellular carcinoma. Cancer Res. 57, 4249–4256 (1997).
World Cancer Research Fund/American Institute for Cancer Research. Food, Nutrition, Physical Activity, and the Prevention of Cancer: a Global Perspective (AICR, Washington, DC, 2007).
Hoeflich, A. et al. Insulin-like growth factor-binding protein 2 in tumorigenesis: protector or promoter? Cancer Res. 61, 8601–8610 (2001).
Fatayerji, N., Engelmann, G.L., Myers, T. & Handa, R.J. In utero exposure to ethanol alters mRNA for insulin-like growth factors and insulin-like growth factor-binding proteins in placenta and lung of fetal rats. Alcohol. Clin. Exp. Res. 20, 94–100 (1996).
Edenberg, H.J. et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res. 34, 840–852 (2010).
Gao, Y. et al. Risk factors for esophageal and gastric cancers in Shanxi Province, China: a case-control study. Cancer Epidemiol. 35, e91–e99 (2011).
He, Z. et al. Prevalence and risk factors for esophageal squamous cell cancer and precursor lesions in Anyang, China: a population-based endoscopic survey. Br. J. Cancer 103, 1085–1088 (2010).
Mark, S.D. et al. Prospective study of serum selenium levels and incident esophageal and gastric cancers. J. Natl. Cancer Inst. 92, 1753–1763 (2000).
Gao, Y. et al. Family history of cancer and risk for esophageal and gastric cancer in Shanxi, China. BMC Cancer 9, 269 (2009).
Lu, S.H. et al. Relevance of N-nitrosamines to esophageal cancer in China. J. Cell Physiol. Suppl. 4, 51–58 (1986).
Acknowledgements
This work was funded by the National High-Tech Research and Development Program of China (2009AA022706 to D.L.), the National Basic Research Program of China (2011CB504303 to D.L. and W.T.), the National Natural Science Foundation of China (30721001 to D.L., Q.Z. and Z.L.) and the Intramural Research Program of the US National Institutes of Health, NCI and the Division of Cancer Epidemiology and Genetics.
Author information
Authors and Affiliations
Contributions
D.L. was the overall principle investigator of the study who conceived the study and obtained financial support, was responsible for study design, oversaw the entire study, interpreted the results and wrote parts of and synthesized the paper. C.W. performed overall project management, oversaw laboratory analyses, performed statistical analyses and drafted the initial manuscript. P.K. oversaw statistical analyses, interpreted the results and reviewed the manuscript. Y. Li and L.L. performed the imputation analysis and reviewed the manuscript. K.Z., J.C., Y.Q., Yuling Zhou and Y. Liu performed laboratory analyses. Z. Hu, G.J. and H.S. were responsible for subject recruitment and sample preparation of Nanjing samples. Z. He, C.G., C.L., H.Y. and Y.K. were responsible for subject recruitment and sample preparation of Henan samples. W.J., J.F. and Y. Zeng were responsible for subject recruitment and sample preparation of Guangzhou samples. X.M. and T.W. provided some of the control samples. Yifeng Zhou was responsible for subject recruitment of the additional validation cohorts. D.Y. and W.T. performed subject recruitment and sample preparation of Beijing samples. Q.Z. and Z.L. provided some of the financial support and reviewed the manuscript. Z.W., C.C.A., N.H., N.D.F., T.D., A.M.G., S.J.C. and P.R.T. performed subject recruitment, sample preparation, laboratory analysis and statistical analysis of Shanxi samples and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–7 and Supplementary Figure 1. (PDF 284 kb)
Rights and permissions
About this article
Cite this article
Wu, C., Kraft, P., Zhai, K. et al. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet 44, 1090–1097 (2012). https://doi.org/10.1038/ng.2411
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2411
This article is cited by
-
HEATR3 involved in the cell proliferation, metastasis and cell cycle development of bladder cancer acts as a tumor suppressor
Molecular Genetics and Genomics (2023)
-
Recent advances in the mechanisms of development and the early diagnosis and treatment of esophageal cancer
Holistic Integrative Oncology (2023)
-
From molecular mechanisms of prostate cancer to translational applications: based on multi-omics fusion analysis and intelligent medicine
Health Information Science and Systems (2023)
-
Global burden and epidemiology of Barrett oesophagus and oesophageal cancer
Nature Reviews Gastroenterology & Hepatology (2021)
-
Oncogenic SNORD12B activates the AKT-mTOR-4EBP1 signaling in esophageal squamous cell carcinoma via nucleus partitioning of PP-1α
Oncogene (2021)