Abstract
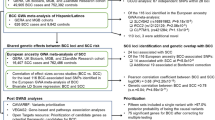
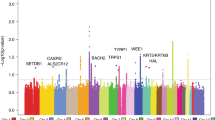
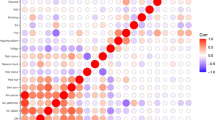
To search for new sequence variants that confer risk of cutaneous basal cell carcinoma (BCC), we conducted a genome-wide SNP association study of 930 Icelanders with BCC and 33,117 controls. After analyzing 304,083 SNPs, we observed signals from loci at 1p36 and 1q42, and replicated these associations in additional sample sets from Iceland and Eastern Europe. Overall, the most significant signals were from rs7538876 on 1p36 (OR = 1.28, P = 4.4 × 10−12) and rs801114 on 1q42 (OR = 1.28, P = 5.9 × 10−12). The 1p36 locus contains the candidate genes PADI4, PADI6, RCC2 and ARHGEF10L, and the gene nearest to the 1q42 locus is the ras-homolog RHOU. Neither locus was associated with fair pigmentation traits that are known risk factors for BCC, and no risk was observed for melanoma. Approximately 1.6% of individuals of European ancestry are homozygous for both variants, and their estimated risk of BCC is 2.68 times that of noncarriers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lear, J.T., Hoban, P., Strange, R.C. & Fryer, A.A. Basal cell carcinoma: from host response and polymorphic variants to tumour suppressor genes. Clin. Exp. Dermatol. 30, 49–55 (2005).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Kutyavin, I.V. et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 34, e128 (2006).
Scherer, D. et al. MC1R variants associated susceptibility to basal cell carcinoma of skin: interaction with host factors and XRCC3 polymorphism. Int. J. Cancer 122, 1787–1793 (2007).
Roewert-Huber, J., Lange-Asschenfeldt, B., Stockfleth, E. & Kerl, H. Epidemiology and aetiology of basal cell carcinoma. Br. J. Dermatol. 157 (Suppl. 2), 47–51 (2007).
Xu, L.Y. & Koo, J. Predictive value of phenotypic variables for skin cancer: risk assessment beyond skin typing. Int. J. Dermatol. 45, 1275–1283 (2006).
Bastiaens, M.T. et al. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. Am. J. Hum. Genet. 68, 884–894 (2001).
Han, J., Colditz, G.A. & Hunter, D.J. Risk factors for skin cancers: a nested case-control study within the Nurses' Health Study. Int. J. Epidemiol. 35, 1514–1521 (2006).
Han, J., Kraft, P., Colditz, G.A., Wong, J. & Hunter, D.J. Melanocortin 1 receptor variants and skin cancer risk. Int. J. Cancer 119, 1976–1984 (2006).
Gudbjartsson, D.F. et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 40, 886–891 (2008).
Sulem, P. et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 39, 1443–1452 (2007).
Sulem, P. et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 40, 835–837 (2008).
Markovic, S.N. et al. Malignant melanoma in the 21st century, part 1: epidemiology, risk factors, screening, prevention, and diagnosis. Mayo Clin. Proc. 82, 364–380 (2007).
Gyorgy, B., Toth, E., Tarcsa, E., Falus, A. & Buzas, E.I. Citrullination: a posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 38, 1662–1677 (2006).
Chavanas, S. et al. Peptidylarginine deiminases and deimination in biology and pathology: relevance to skin homeostasis. J. Dermatol. Sci. 44, 63–72 (2006).
Suzuki, A., Yamada, R. & Yamamoto, K. Citrullination by peptidylarginine deiminase in rheumatoid arthritis. Ann. NY Acad. Sci. 1108, 323–339 (2007).
Wysocka, J., Allis, C.D. & Coonrod, S. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11, 344–355 (2006).
Esposito, G. et al. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol. Cell. Endocrinol. 273, 25–31 (2007).
Mollinari, C. et al. The mammalian passenger protein TD-60 is an RCC1 family member with an essential role in prometaphase to metaphase progression. Dev. Cell 5, 295–307 (2003).
Winkler, S., Mohl, M., Wieland, T. & Lutz, S. GrinchGEF—a novel Rho-specific guanine nucleotide exchange factor. Biochem. Biophys. Res. Commun. 335, 1280–1286 (2005).
Tao, W., Pennica, D., Xu, L., Kalejta, R.F. & Levine, A.J. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 15, 1796–1807 (2001).
Yang, S.H. et al. Pathological responses to oncogenic Hedgehog signaling in skin are dependent on canonical Wnt/β-catenin signaling. Nat. Genet. 40, 1130–1135 (2008).
Hahn, H. et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85, 841–851 (1996).
Johnson, R.L. et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671 (1996).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008).
O'Driscoll, L. et al. Investigation of the molecular profile of basal cell carcinoma using whole genome microarrays. Mol. Cancer 5, 74 (2006).
Barrett, J.C. & Cardon, L.R. Evaluating coverage of genome-wide association studies. Nat. Genet. 38, 659–662 (2006).
Gretarsdottir, S. et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 35, 131–138 (2003).
Acknowledgements
We thank G.H. Olafsdottir for assistance in the ascertainment of affected individuals.
Author information
Authors and Affiliations
Contributions
The study was designed and results were interpreted by S.N.S., D.F.G., P.S., A.K., J.H.O., U.T. and K.S. Statistical analysis was carried out by D.F.G., P.S., G.T., A.H., F.G. and A.K. Subject ascertainment and recruitment was organized and carried out by S.N.S., R.K., B.S., K.R.B., K.T., R.R., D.S., P.R., E.G., K. Koppova, V.M., R.B.-E., V.S., P.J., M.G., F.J.C., P.T., Y.G., K. Kristjansson, G.B., S.G.S., E.N., J.I.M., J.H., J.H.O. and U.T. Biological material collection and handling was supervised by S.N.S., D.S., P.R., E.G., K. Koppova, V.M., R.B.-E., V.S., P.J., M.G., F.J.C., P.T., Y.G., T.R., M.J., J.G., S.T., J.S., K. Kristjansson, S.G.S., E.N., J.I.M., J.H. and U.T. Genotyping was supervised by S.N.S., T.R., M.J., J.G., S.T., M.M. and U.T. Expression analysis was carried out by J.T.B., G.T., A.S., T.T., A.J. and T.B. Bioinformatic analysis was carried out by S.A.G. and G.F.J. Authors S.N.S., D.F.G., P.S., U.T. and K.S. drafted the manuscript. All authors contributed to the final version of the paper. Principal collaborators for the case-control population samples were J.H.O. (Iceland), R.K. (Eastern Europe), J.I.M. and E.N. (Spain), and J.H. (Sweden).
Corresponding authors
Ethics declarations
Competing interests
Authors whose affiliations are listed as deCODE Genetics are shareholders and/or employees of deCODE Genetics, a biotechnology company. deCODE Genetics intends to incorporate the variants described in this paper into its genetic-testing services.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–3 and Supplementary Figures 1 and 2 (PDF 159 kb)
Rights and permissions
About this article
Cite this article
Stacey, S., Gudbjartsson, D., Sulem, P. et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet 40, 1313–1318 (2008). https://doi.org/10.1038/ng.234
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.234
This article is cited by
-
Understanding the matrix: collagen modifications in tumors and their implications for immunotherapy
Journal of Translational Medicine (2024)
-
Multi-ancestry genome-wide meta-analysis identifies novel basal cell carcinoma loci and shared genetic effects with squamous cell carcinoma
Communications Biology (2024)
-
A novel tumor suppressor encoded by a 1p36.3 lncRNA functions as a phosphoinositide-binding protein repressing AKT phosphorylation/activation and promoting autophagy
Cell Death & Differentiation (2023)
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
A systematic review of skin ageing genes: gene pleiotropy and genes on the chromosomal band 16q24.3 may drive skin ageing
Scientific Reports (2022)