Abstract
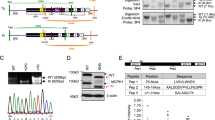
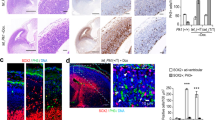
Large brain size is one of the defining characteristics of modern humans. Seckel syndrome (MIM 210600), a disorder of markedly reduced brain and body size1,2, is associated with defective ATR-dependent DNA damage signaling3. Only a single hypomorphic mutation of ATR has been identified in this genetically heterogeneous condition4. We now report that mutations in the gene encoding pericentrin (PCNT)—resulting in the loss of pericentrin from the centrosome, where it has key functions anchoring both structural and regulatory proteins—also cause Seckel syndrome5,6. Furthermore, we find that cells of individuals with Seckel syndrome due to mutations in PCNT (PCNT-Seckel) have defects in ATR-dependent checkpoint signaling, providing the first evidence linking a structural centrosomal protein with DNA damage signaling. These findings also suggest that other known microcephaly genes implicated in either DNA repair responses7 or centrosomal function8,9 may act in common developmental pathways determining human brain and body size.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Majewski, F. et al. Studies of microcephalic primordial dwarfism I: approach to a delineation of the Seckel syndrome. Am. J. Med. Genet. 12, 7–21 (1982).
Seckel, H.P.G. Bird-headed Dwarfs: Studies in Developmental Anthropology Including Human Proportions (Charles C. Thomas, Springfield, Illinois, 1960).
Alderton, G.K. et al. Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum. Mol. Genet. 13, 3127–3138 (2004).
O'Driscoll, M. et al. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33, 497–501 (2003).
Diviani, D. et al. Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr. Biol. 10, 417–420 (2000).
Zimmerman, W.C. et al. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15, 3642–3657 (2004).
Alderton, G.K. et al. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat. Cell Biol. 8, 725–733 (2006).
Bond, J. et al. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 32, 316–320 (2002).
Bond, J. et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37, 353–355 (2005).
Shiloh, Y. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11, 71–77 (2001).
Flory, M.R. et al. The centrosomal proteins pericentrin and kendrin are encoded by alternatively spliced products of one gene. Genomics 82, 401–405 (2003).
Miyoshi, K. et al. Characterization of pericentrin isoforms in vivo. Biochem. Biophys. Res. Commun. 351, 745–749 (2006).
Li, Q. et al. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J. Cell Sci. 114, 797–809 (2001).
Dictenberg, J.B. et al. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141, 163–174 (1998).
Doxsey, S.J. et al. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76, 639–650 (1994).
Purohit, A. et al. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147, 481–492 (1999).
Martinez-Campos, M. et al. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673–683 (2004).
Chen, D. et al. Centrosomal anchoring of protein kinase C betaII by pericentrin controls microtubule organization, spindle function, and cytokinesis. J. Biol. Chem. 279, 4829–4839 (2004).
Sengupta, S. et al. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J. Cell Biol. 166, 801–813 (2004).
Woods, C.G. et al. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am. J. Hum. Genet. 76, 717–728 (2005).
Ponting, C. et al. Evolution of primary microcephaly genes and the enlargement of primate brains. Curr. Opin. Genet. Dev. 15, 241–248 (2005).
Brown, P. et al. A new small-bodied hominin from the Late Pleistocene of Flores, Indonesia. Nature 431, 1055–1061 (2004).
Martin, R.D. et al. Flores hominid: new species or microcephalic dwarf? Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288, 1123–1145 (2006).
Doxsey, S. et al. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21, 411–434 (2005).
Zhang, S. et al. Centrosomal localization of DNA damage checkpoint proteins. J. Cell. Biochem. 101, 451–465 (2007).
Loffler, H. et al. DNA damage–induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle 6, 2541–2518 (2007).
Kramer, A. et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 6, 884–891 (2004).
Niida, H. et al. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol. Cell. Biol. 27, 2572–2581 (2007).
Ruschendorf, F. et al. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics 21, 2123–2125 (2005).
Dodson, H. et al. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 23, 3864–3873 (2004).
Acknowledgements
We thank the families and their clinicians for their participation in this study; C. Hayward for contributing control samples; S. McKay and the MRC HGU core sequencing service for advice and technical support; C. Nicol for help with figure preparation; X. Fant, V. Van Heyningen and N. Hastie for discussions and comments; W. Fergusson (St. Mary's Hospital, Manchester) for LCL transformation; and A. Merdes and J. Salisbury for kindly sharing antibody reagents. A.P.J.'s laboratory is funded by the MRC, M.O'D.'s laboratory is funded by CRUK and the MRC, and P.A.J.'s laboratory is funded by the MRC, UK LRF, IACR and EU grants (FIGH-CT-200200207) (DNA repair) and FI6R-CT-2003-508842 (RiscRad). P.V. and W.C.E. are funded by the Wellcome Trust, of which W.C.E. is a Principal Research Fellow. A.P.J. is an MRC Senior Clinical Fellow and M.O'D. is a CRUK Senior Cancer Research Fellow.
Author information
Authors and Affiliations
Contributions
E.G., C.-A.M. and A.P.J. performed the mutation screening and sequencing of controls; A.P.J., linkage analysis; P.V., immunostaining analysis of LCLs and C.-A.M., immunoblotting. S.W., T.S., M.O'D. and P.A.J. designed and performed the DNA damage response assays and RNAi experiments. N.A.S., A.S. and B.H. provided clinical samples and data. E.G. and A.P.J. wrote the paper with M.O'D., P.A.J., B.V. and W.C.E.
Corresponding author
Supplementary information
Supplementary Figures and Text
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 615 kb)
Rights and permissions
About this article
Cite this article
Griffith, E., Walker, S., Martin, CA. et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet 40, 232–236 (2008). https://doi.org/10.1038/ng.2007.80
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2007.80
This article is cited by
-
An updated view on the centrosome as a cell cycle regulator
Cell Division (2022)
-
Centrosome, microtubule and DNA damage response
Genome Instability & Disease (2022)
-
Microcephalic osteodysplastic primordial dwarfism type II is associated with global vascular disease
Orphanet Journal of Rare Diseases (2021)
-
Identification of three novel mutations in PCNT in vietnamese patients with microcephalic osteodysplastic primordial dwarfism type II
Genes & Genomics (2021)
-
Loss of the centrosomal protein Cenpj leads to dysfunction of the hypothalamus and obesity in mice
Science China Life Sciences (2021)