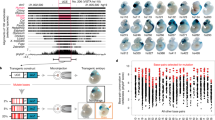
Abstract
Extended perfect human-rodent sequence identity of at least 200 base pairs (ultraconservation) is potentially indicative of evolutionary or functional uniqueness. We used a transgenic mouse assay to compare the embryonic enhancer activity of 231 noncoding ultraconserved human genome regions with that of 206 extremely conserved regions lacking ultraconservation. Developmental enhancers were equally prevalent in both populations, suggesting instead that ultraconservation identifies a small, functionally indistinct subset of similarly constrained cis-regulatory elements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Waterston, R.H. et al. Nature 420, 520–562 (2002).
Bejerano, G. et al. Science 304, 1321–1325 (2004).
Katzman, S. et al. Science 317, 915 (2007).
Derti, A., Roth, F.P., Church, G.M. & Wu, C.T. Nat. Genet. 38, 1216–1220 (2006).
Drake, J.A. et al. Nat. Genet. 38, 223–227 (2006).
Cooper, G.M. et al. Genome Res. 15, 901–913 (2005).
Siepel, A. et al. Genome Res. 15, 1034–1050 (2005).
de la Calle-Mustienes, E. et al. Genome Res. 15, 1061–1072 (2005).
Poulin, F. et al. Genomics 85, 774–781 (2005).
Pennacchio, L.A. et al. Nature 444, 499–502 (2006).
Ghanem, N. et al. J. Neurosci. 27, 5012–5022 (2007).
Paparidis, Z. et al. Dev. Growth Differ. 49, 543–553 (2007).
Prabhakar, S. et al. Genome Res. 16, 855–863 (2006).
Hardison, R.C., Oeltjen, J. & Miller, W. Genome Res. 7, 959–966 (1997).
Ahituv, N. et al. PLoS Biol. 5, e234 (2007).
Acknowledgements
We thank I. Dubchak, A. Poliakov and S. Minovitsky for help with genome alignments and database development; S. Bhardwaj and S. Phouanenavong for technical assistance; and N. Ahituv, M. Nobrega, J. Noonan and members of the Pennacchio and Rubin laboratories for discussion and critical comments on the manuscript. L.A.P. was supported by grant HL066681, Berkeley-Program for Genomic Applications, under the Programs for Genomic Applications, funded by National Heart, Lung, and Blood Institute, and HG003988 funded by National Human Genome Research Institute. Research was done under Department of Energy Contract DE-AC02-05CH11231, University of California, E.O. Lawrence Berkeley National Laboratory. A.V. was supported by an American Heart Association postdoctoral fellowship.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Tables 1,3 and 5, Supplementary Figures 1 and 2 (PDF 315 kb)
Supplementary Table 2
Genome-wide set of extremely human-rodent constrained non-coding sequences (XLS 284 kb)
Supplementary Table 4
Genome-wide enhancer testing of non-exonic ultraconserved elements (XLS 58 kb)
Rights and permissions
About this article
Cite this article
Visel, A., Prabhakar, S., Akiyama, J. et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40, 158–160 (2008). https://doi.org/10.1038/ng.2007.55
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2007.55
This article is cited by
-
Genomic insights into the seawater adaptation in Cyprinidae
BMC Biology (2024)
-
A distant global control region is essential for normal expression of anterior HOXA genes during mouse and human craniofacial development
Nature Communications (2024)
-
Evolutionary relevance of single nucleotide variants within the forebrain exclusive human accelerated enhancer regions
BMC Molecular and Cell Biology (2023)
-
Decoding enhancer complexity with machine learning and high-throughput discovery
Genome Biology (2023)
-
Using Synthetic DNA Libraries to Investigate Chromatin and Gene Regulation
Chromosoma (2023)