Abstract
Replicative stress during embryonic development influences ageing and predisposition to disease in adults. A protective mechanism against replicative stress is provided by the licensing of thousands of origins in G1 that are not necessarily activated in the subsequent S-phase. These ‘dormant’ origins provide a backup in the presence of stalled forks and may confer flexibility to the replication program in specific cell types during differentiation, a role that has remained unexplored. Here we show, using a mouse strain with hypomorphic expression of the origin licensing factor mini-chromosome maintenance (MCM)3 that limiting origin licensing in vivo affects the functionality of hematopoietic stem cells and the differentiation of rapidly-dividing erythrocyte precursors. Mcm3-deficient erythroblasts display aberrant DNA replication patterns and fail to complete maturation, causing lethal anemia. Our results indicate that hematopoietic progenitors are particularly sensitive to replication stress, and full origin licensing ensures their correct differentiation and functionality.
Similar content being viewed by others
Introduction
The process of genomic duplication starts at replication origins, which are licensed in the G1 phase of the cell division cycle, several hours before their activation in S phase. The licensing process is led by the origin recognition complex (ORC), cell division cycle 6 (CDC6) and Cdc10-dependent transcript 1 (CDT1) proteins, which cooperate to engage the mini-chromosome maintenance (MCM) complex with the DNA. MCM, composed by essential subunits MCM2-7, displays DNA helicase activity and becomes part of the replisome machinery (reviewed in references1,2). Defective control of DNA replication causes ‘replicative stress’ (RS), which is the underlying cause of several developmental diseases. Mutations in ORC, CDC6 and CDT1 genes are related to Meier-Gorlin syndrome, a type of dwarfism3,4,5, and mutations in MCM4 are linked to growth retardation, adrenal insufficiency and natural killer cell deficiency6. Impaired MCM function also increases cancer susceptibility7,8,9,10,11,12,13 (reviewed in references14,15).
MCM complexes are normally loaded onto DNA in excess relative to the number of origins that fire during the S phase (reviewed in reference16). One function of the surplus of MCM is to license dormant origins that may be activated in response to stalled or collapsed forks, providing a rescue mechanism under RS17,18,19. Another possible function for the high number of licensed origins, which remains largely underexplored, is to provide flexibility to the replication process during early embryonic development20,21 or in cell differentiation contexts that require the activation or shut-off of specific origins22,23,24 (reviewed in references25,26).
To investigate the protective effects of MCM against RS in vivo, a new Mcm3 conditional knockout (KO) mouse model was designed. The targeted allele, Mcm3-Lox, was hypomorphically expressed and caused embryonic lethality in homozygosity, which could be partially alleviated in a genetic background with enhanced resistance to RS. Our results show how a full complement of MCM proteins is specifically required to preserve the functionality of hematopoietic stem cells (HSCs) and the proper differentiation and maturation of erythrocytes in the developing embryo.
Results
A hypomorphic Mcm3 allele causes embryonic lethality
A modified mouse Mcm3 allele was designed with loxP sites flanking exons 14–17 and a luciferase reporter inserted at the 3′ UTR under the control of an IRES element. The resultant allele (Mcm3-Lox) was intended as a conditional KO, as Mcm3 expression could be ablated with Cre recombinase (Fig. 1a and Supplementary Fig. 1A,B). In addition, expression of Mcm3-Lox could be monitored by the bioluminescence activity associated to luciferase expression. Mcm3+/Lox mice were born at Mendelian rates, and Mcm3-Lox expression in the skin and several internal organs was confirmed by luciferase activity after the administration of luciferin (Fig. 1b and Supplementary Fig. 1C). Unexpectedly, virtually no Mcm3Lox/Lox individuals were born after extensive breeding of Mcm3+/Lox mice (1 in 314; Fig. 1c). Mcm3Lox/Lox embryos survived until E16.5-E18.5 but were noticeably smaller than wild-type (wt) or Mcm3+/Lox littermates and their pale appearance suggested an anemic phenotype (Fig. 1d). Mcm3+/Lox and Mcm3Lox/Lox embryonic fibroblasts (MEFs) could be obtained and maintained in culture.
(a) Schematic of the endogenous Mcm3 locus (Mcm3+) and the different alleles resulting from targeted recombination. Mcm3Lox-Neo carries loxP sites flanking exons 14 and 17, a luciferase reporter expressed from an IRES element, and a neomycin-resistance cassette flanked by frt sites (gray triangles) that can be excised with FlpE recombinase, resulting in the Mcm3Lox allele. Cre recombinase excises exons 14–17, resulting in the Mcm3− (null) allele. (b) Imaging of whole body bioluminescence of Mcm3+/+ and Mcm3+/Lox mice, taken 10 min after intraperitoneal injection of luciferin. Mice were partially shaved on the back to expose the skin. (c) Percentage (expected and obtained) of Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox mice derived from extensive Mcm3+/Lox × Mcm3+/Lox breeding. (d) Representative images of Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox E16.5 embryos and placental tissue. Scale bar, 1 cm. (e) Mcm3 mRNA levels in Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox MEFs, determined by qRT-PCR. Histogram shows the average ±s.d. of 3 independent experiments. P-values were calculated with Fisher’s test (***P<0.001). (f) Protein levels of the indicated MCM subunits in Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox MEFs as determined by immunoblots. For accurate comparisons, different amounts of each extract were loaded. For each SDS-PAGE, MEK2 levels are shown as loading control. Full western blots are shown in Supplementary Fig. 9A. (g) Percentage (expected and obtained) of Mcm3+/+, Mcm3+/− and Mcm3−/− mice after extensive Mcm3+/− × Mcm3+/− breeding. (h) Kaplan-Meier survival curves for Mcm3+/+ (black), Mcm3+/Lox (red) and Mcm3+/− (blue) mice. 42 individuals were included in the wt group (18 female; 24 male), 31 individuals in the +/Lox group (15 female; 16 male) and 35 individuals in the +/- group (13 female; 22 male). Log-Rank (Mantel-Cox) test indicated that there were significant differences between Mcm3+/+ and Mcm3+/Lox or Mcm3+/− curves (***P<0.001).
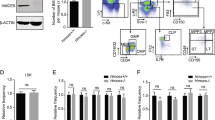
To determine the impact of the Mcm3-Lox modification on gene function, mRNA isolated from MCM3Lox/Lox MEFs was sequenced to rule out the presence of any mutation that could affect MCM3 protein functionality (Supplementary Fig. 2). Next, Mcm3 expression was compared in MEFs derived from Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox embryos. Mcm3 mRNA levels were reduced to approximately 70% in Mcm3+/Lox and 30% in Mcm3Lox/Lox MEFs, relative to the levels in Mcm3+/+ MEFs (Fig. 1e), and consistent reductions in MCM3 protein were observed in whole cell extracts (Fig. 1f; compare lanes 3–5–7). These results indicate that the Mcm3-Lox allele is hypomorphically expressed, presumably due to the modifications introduced in the 3′ UTR. This level of reduction in MCM3 did not significantly affect the cellular concentration of other MCM subunits (Fig. 1f).
At least in primary embryonic fibroblasts, downregulation of Mcm3 expression was compatible with DNA replication and cell proliferation (Supplementary Fig. 3A,B). Analyses of inter-origin distances (IOD) using stretched fibers revealed a similar frequency of origin activity between Mcm3+/+ and Mcm3Lox/Lox MEFs in normal growth conditions (median IOD values of 118 and 116 Kb, respectively; Supplementary Fig. 3C). Upon challenging fork progression with aphidicolin, the IOD was shortened by 37% in Mcm3+/+ MEFs, as a consequence of the activation of dormant origins (median IOD 73.8 Kb; Supplementary Fig. 3C). In Mcm3Lox/Lox MEFs, the reduction in IOD was limited to 24% (median IOD 87.5 Kb), reflecting a partial loss of back-up origins but also underscoring the large excess of origins licensed in G1, as even after the marked reduction in MCM3 protein concentration, a reservoir of additional origins were activated when needed. After several days in culture, Mcm3Lox/Lox MEFs displayed γH2AX foci, a common marker of DNA damage associated to RS (Supplementary Fig. 3D).
Mcm3-deficient mice are prone to haematological neoplasia
In contrast to Mcm3Lox/Lox, heterozygous Mcm3+/Lox mice were viable and could be crossed with a CMV-Cre strain expressing Cre recombinase to generate viable Mcm3+/− mice. As expected, breeding between Mcm3+/− mice did not produce any Mcm3−/− offspring (Fig. 1g). All previous attempts at making MCM-null models displayed pre-implantation lethality8,9. When cohorts of Mcm3+/− and Mcm3+/Lox mice were established for longevity studies, both groups presented a slight but significant reduction in lifespan (Fig. 1h) and an increased incidence of tumors, mainly lymphomas originated in the mesenteric lymph nodes that in multiple cases infiltrated to other organs. These tumors are not uncommon in aged mice but normally have a later onset and lower invasiveness (Supplementary Tables 1–3).
DNA damage and incomplete erythropoiesis in the fetal liver
We next focused on the tissues that might be responsible for the lethality observed in Mcm3Lox/Lox embryos, which by mid-late gestation (E14.5–18.5) were consistently smaller than their Mcm3+/+ or Mcm3+/Lox counterparts. In control E16.5 embryos, MCM3 protein is readily detected by immunohistochemistry (IHC) in most tissues, particularly liver, lung, thymus, brown adipose tissue and the subventricular zone of the brain. In Mcm3Lox/Lox embryos, the strong MCM3 staining in the liver was markedly reduced while γH2AX staining became apparent (Fig. 2). Mcm3 mRNA and protein levels confirmed the downregulation of Mcm3 expression in fetal liver extracts (Supplementary Fig. 4A,B). Global levels of cell proliferation were not affected, as monitored by Ki67 IHC staining (Supplementary Fig. 4C).
(a) Immunohistochemistry (IHC) detection of MCM3 and γH2AX proteins in Mcm3+/+ and Mcm3Lox/Lox embryo (E16.5) sections. The following tissues are indicated: SVZ, subventricular zone of the brain; Thy, thymus; BAT, brown adipose tissue; lung; liver. Scale bar, 2 mm. (b) Detailed sections of the indicated tissues stained for MCM3 and γH2AX. Scale bars: SVZ, 200 μm; thymus, 20 μm; BAT, 100 μm; lung, 50 μm; liver, 50 μm.
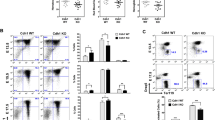
The liver is the major site of fetal hematopoiesis from E11 until shortly before birth, when this process becomes established in the bone marrow. To evaluate the impact of Mcm3 downregulation in hematopoiesis, specific blood lineages were detected by IHC in mid-late gestation embryos. This approach did not reveal major differences in the abundance of mature B-lymphocytes (stained with Pax-5) and megakaryocytes (stained with FVIII) in the fetal liver, nor T-lymphocytes (stained with CD3) in the fetal thymus of Mcm3Lox/Lox embryos compared to their Mcm3+/+ or Mcm3+/Lox counterparts (Supplementary Fig. 5). In contrast, the staining with erythrocyte marker Ter119 was much reduced in the liver of Mcm3Lox/Lox embryos (Fig. 3a). Peripheral blood from Mcm3-deficient E14.5 embryos displayed a marked decrease in red blood cells (RBCs), haemoglobin concentration and hematocrit (Fig. 3b), all consistent with a phenotype of fetal anemia. Giemsa staining of peripheral blood cells revealed a large accumulation of immature erythroblasts (EBs) with uncompacted nuclei, as well as the presence of micronuclei that are indicative of genomic instability (Fig. 3c,d).
(a) Ter119 IHC staining in Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox E16.5 embryos. Scale bar, 5 mm. (b) Peripheral blood counts from Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox E14.5 embryos. RBC, number of red blood cells; HGB, haemoglobin concentration; HCT, hematocrit. Histograms show the average value±s.d. of Mcm3+/+ (n=6), Mcm3+/Lox (n=7) and Mcm3Lox/Lox (n=3) embryo peripheral blood cell counts. P-values were calculated by Fisher’s test (***P<0.001, **P<0.05, NS, not significant). (c) Representative fields of Giemsa-stained peripheral blood samples derived from Mcm3+/+ and Mcm3Lox/Lox embryos. Scale bar, 20 μm. (d) Percentage of nucleated erythrocytes in embryo peripheral blood samples. Histograms show the average value±s.d. of Mcm3+/+ (n=5), Mcm3+/Lox (n=8) and Mcm3Lox/Lox (n=6) cell counts. Statistical significance was calculated by One-way Anova (**P<0.05, NS, not significant). (e) Top, schematic of the pathway of erythrocyte differentiation. Bottom left, density plots showing the distribution of EBs in R1-R5 stages in Mcm3+/+ and Mcm3Lox/Lox fetal livers. Bottom right, histogram quantification of the percentage (±s.d.) of cells in R1-R5 in Mcm3+/+ (n=15) and Mcm3Lox/Lox (n=7) fetal livers. P-values were calculated by Fisher’s test (***P<0.001, NS not significant).
Erythrocyte differentiation is coupled to 3–5 rounds of DNA replication and cell division27. Based on the expression of surface markers CD71 and Ter119, EBs can be separated by flow cytometry in consecutive differentiation stages28: pro-EBs (located in ‘region’ R1); early basophilic EBs (baso-EBs; R2); early and late baso-EBs (R3); chromatophilic/orthochromatophilic EBs (chr-EBs/ orth-EBs; R4); and late orth-EBs/ reticulocytes (R5). In agreement with the analyses of embryo peripheral blood, the fetal liver of Mcm3Lox/Lox embryos displayed an accumulation of pro-EBs and early baso-EBs, and a concomitant reduction in chr-EBs, orth-EBs and mature reticulocytes (Fig. 3e). Activated caspase 3 was detected in a fraction of R3 Mcm3-deficient EBs, indicating apoptotic events (Supplementary Fig. 6).
Impaired erythrocyte maturation upon transplantation
The fetal liver contains HSCs capable of reconstituting the entire hematopoietic system. In order to compare the maturation ability of wild-type and Mcm3-deficient cells towards the erythroid lineage, fetal liver cells (E14.5) derived from wild-type or Mcm3-deficient embryos were transplanted into lethally-irradiated recipient mice, in competition (1:1 ratio) with bone marrow cells that constitutively express a Tomato (Tom) fluorescent marker29 (Fig. 4a). Two months after transplantation, the contribution of control Mcm3+/+ cells (Tom-negative) towards mature RBCs in peripheral blood was approximately 50% in the chimaeras (Fig. 4b). In contrast, the contribution of Mcm3-deficient cells (Tom-negative) towards RBCs was much reduced, with Mcm3-proficient cells (Tom-positive) taking over most of the RBC population (Fig. 4b). These results confirm that EBs with reduced expression of Mcm3 do not mature properly under steady-state conditions.
(a) Experimental design of a competitive transplantation assay between Mcm3+/+ and Mcm3Lox/Lox fetal liver (FL; E14.5) cells and bone marrow (BM) cells of a tool strain constitutively expressing red fluorescent Tomato (Tom) protein, with wild-type Mcm3 levels. In each case, cells were transplanted at 1:1 ratio. (b) Quantification of the RBC (Ter119-positive) chimerism in peripheral blood of recipient mice, 2 months post-transplantation. Histograms show average±s.d.; n=4 mice transplanted with Mcm3+/+ cells and n=4 mice transplanted with Mcm3Lox/Lox cells. P-values were calculated by Fisher’s test (***P<0.001, NS, not significant). (c) Schematic of a phlebotomy assay in chimaeric mice to monitor the erythropoietic stress response. (d) Examples of flow cytometry analyses of splenic EryA, EryB, EryC precursors (abbreviated A, B, C), in control mice (CTR) or mice subjected to phlebotomy (PHL). Analyses of EryA, EryB and EryC was carried out four days after the bleeding, differentiating between Tom+ and Tom- cells. (e) Left, histograms showing the percentage of EryA, EryB and EryC populations relative to control mice (n= 2 CTR, n=4 PHL mice). P-values were calculated by Fisher’s test (***P<0.001, **P<0.01). Right, histogram showing the fold-change variation in Tom+ (red) or Tom- (white) EryA, EryB and EryC populations induced by phlebotomy.
Chimaeric Mcm3+/+ (Tom+) / Mcm3Lox/Lox (Tom-) mice were also used to monitor the acute response to erythropoietic stress induced by phlebotomy (Fig. 4c). In this setting, cells in the spleen with Ter119 high content can be analyzed using the forward scatter parameter (FSC) and CD71 to resolve three EB subpopulations previously labelled as EryA, EryB and EryC30. EryA (Ter119high CD71high FSChigh) correspond to baso-EBs; EryB (Ter119high CD71high FSClow) correspond to late baso-EBs and polychromatic-EBs; EryC (Ter119high CD71low FSClow) are orth-EBs and reticulocytes. In response to stress, the EryA and EryB subpopulations are rapidly increased30. Both Mcm3-competent and Mcm3-deficient EBs reacted to stress, as indicated by the accumulation of splenic EryA and EryB progenitors (Fig. 4d,e). However, the accumulation of EryA in Mcm3-deficient cells was almost 3-fold higher than in Mcm3+/+ cells, suggesting that the rapid RBC maturation required under stress conditions is also delayed or impaired (Fig. 4e, right).
Aberrant DNA replication during erythrocyte maturation
To monitor whether inefficient erythrocyte maturation was related to DNA replication and cell cycle progression, fetal liver cells obtained from Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox embryos were sorted at the R1-R4 stages (R5 was not included as most cells have started the enucleation process). Interestingly, Mcm3Lox/Lox baso-EBs (R3) and chr-EBs (R4) displayed a prominent accumulation in S phase, indicative of problems to complete DNA replication (Fig. 5a). High levels of γH2AX and a higher frequency of fork asymmetry (indicative of stalled forks) were detected in Mcm3Lox/Lox early EBs, consistent with RS interfering with RBC maturation (Fig. 5b,c).
(a) DNA content of EBs in R1-R4 differentiation stages. Three examples of each genotype are shown. (b) γH2AX intensity in Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox R3 EBs (>300 nuclei scored per condition). Data are representative of 3 independent experiments. P-values were calculated by Mann-Whitney test (***P<0.001). (c) Box plots showing the values of fork asymmetry in Mcm3+/+ and Mcm3Lox/Lox EBs sorted in R1 and R3 stages. >180 bidirectional forks were counted per condition. Statistical analysis was done with Mann-Whitney rank sum test (***P<0.001). (d) Left, IOD values (data distribution and median value) of Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox EBs at the indicated (R1-R4) maturation stage. >100 IOD values were scored for each condition. P-values were calculated using Mann-Whitney test (***P<0.001; NS, not significant). Right, origin density expressed by number of origins/Mb of DNA was estimated as indicated in Materials and Methods. See also Table 1.
Next, we analyzed the patterns of origin activity in EB populations sorted from embryonic livers using stretched DNA fibers labelled with thymidine analogs chloro-deoxyuridine (CldU) and iodo-deoxyuridine (IdU). These analyses revealed a dynamic pattern in which more origins are activated as EBs progress in the differentiation pathway. The median IOD was progressively reduced from approximately 100 Kb in pro-EB (R1) to 74 Kb in early baso-EBs (R2), 69 Kb in late baso-EBs (R3) and 56 Kb in orth-EBs (R4; Fig. 5d, left). The length of DNA fibers was similar in all populations, ruling out indirect effects on IOD measurements (Supplementary Fig. 7). Furthermore, the tendency towards more origin activity was confirmed by measuring origin density (Fig. 5d, right and Table 1). Interestingly, the patterns of origin activity were altered in Mcm3-deficient EBs, which displayed lower IOD values and slightly higher origin density (Fig. 5d and Table 1). As discussed below, this behaviour likely reflects the increased pressure on early EBs to replicate as a compensatory mechanism for the lack of mature RBCs.
Mcm3Lox/Lox lethality can be rescued by CHK1 overexpression
Because our results strongly point to RS as the underlying cause for incomplete RBC maturation, we considered the possibility that the embryonic lethality of the Mcm3Lox/Lox genotype could be alleviated in a background with higher tolerance to RS, such as the recently described strain carrying an extra copy of checkpoint kinase CHK1 (ref. 31). Mcm3+/Lox:Chk1+/Tg mice were generated by direct intercrossing between both strains, and further breading of Mcm3+/Lox:Chk1+/Tg mice resulted in Mcm3Lox/Lox individuals that successfully completed gestation, despite having low expression levels of Mcm3 (Supplementary Fig. 8). A few Mcm3Lox/Lox individuals were born without the extra Chk1 allele, indicating that the change in genetic background (now mixed C57BL/6-CD1) also contributed to survival of the Mcm3Lox/Lox embryos. Independently of the effect of the mixed background, the birth rate was biased towards Mcm3Lox/Lox: Chk1+/Tg progeny. This partial rescue of embryonic lethality provides further genetic evidence that RS caused by Mcm3 downregulation is the reason for the phenotypes observed in the Mcm3Lox/Lox strain.
Impaired fitness of Mcm3Lox/Lox hematopoietic stem cells
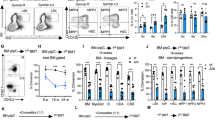
A recent report has linked RS to the limited functionality observed in aged HSCs32. Our Mcm3-deficient model provided a proper genetic tool to validate this concept, as Mcm3-deficient fetal HSCs should also be affected. Fetal Mcm3Lox/Lox LSK cells (Lin−/cKit+/Sca1+), which comprise both HSCs (Lin−/cKit+/Sca1+/Flk2−/CD48−/CD150+) and multipotent progenitor cells (MPPs: Lin−/cKit+/Sca1+/Flk2+), had lower amounts of MCM3 protein and higher levels of RS marker γH2AX (Fig. 6a). Intriguingly, single-cell tracking of cell division revealed slightly faster proliferation kinetics of Mcm3Lox/Lox than Mcm3+/+ fetal HSCs (Fig. 6b). This could reflect a pressure to proliferate and differentiate into MPPs, as it has been described for adult HSCs in the presence of DNA damage33. Consisting with this notion, immunophenotyping analyses revealed a two-fold increase in the concentration of LSK cells in Mcm3Lox/Lox embryos, caused by the accumulation of MPPs (Fig. 6c).
(a) Representative images of LSK cells (Lin−, cKit+, Sca1+) isolated from Mcm3+/+ and Mcm3Lox/Lox embryonic liver, stained with MCM3 (red) and γH2AX (green) antibodies. Scale bar, 5 μM. Right panels, quantification of MCM3 or γH2AX staining intensity, using Definiens software on confocal microscopy images. Red lines indicate the median nuclear intensity (>800 nuclei scored per condition). P-values were calculated by Mann-Whitney test (***P<0.001). (b) Single-cell tracking of the first cell division of cultured fetal Mcm3+/+ or Mcm3Lox/Lox HSCs. Representation shows the average±s.d. of 4 independent experiments. (c) Histograms showing the percentage of LSK, MPP and HSCs in Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox embryonic liver tissue. Median values±s.d. are shown (n=7 embryos for each genotype). P-values were calculated by Fisher’s test (*P<0.05; NS, not significant). (d) HSC engraftment results at 4 months post-transplantation. Middle histograms show the percentage of donor-derived chimerism in the bone marrow (BM), spleen and peripheral blood. Right histograms show the percentage of chimerism in the adult HSC compartment. Quantification shows average±s.d. of three independent experiments. P-values were calculated by Fisher’s test (***P<0.001; *P<0.05; NS, not significant). (e) Confocal microscopy images of γH2AX and RPA staining in donor-derived, cycling adult Mcm3+/+ or Mcm3Lox/Lox HSCs isolated from the BM. Scale bar, 10 μm. (f) Histogram showing the average number of colonies (±s.d.) formed 7 days after plating Mcm3+/+ or Mcm3Lox/Lox HSCs in methylcellulose, in the absence or presence of low-dose aphidicolin (50 ng ml−1). Results are normalized for vehicle-treated cells (DMSO) and expressed as fold change compared to Mcm3+/+ HSCs. 3 independent experiments were conducted and included in the quantification. P-values were calculated using Fisher’s test (*P<0.05; NS, not significant). (g) Donor-derived adult Mcm3+/+ and Mcm3Lox/Lox HSCs were re-isolated and transplanted into lethally irradiated secondary recipients (500 HSCs per mouse; 5 mice per group). Histogram shows the percentage of donor-derived chimerism in peripheral blood, assessed 1–4 months post-transplantation. Data are mean values±s.d. P-values were calculated using Fisher’s test (***P<0.001).
To directly test their functionality, fetal HSCs were isolated from E14.5-E16.5 CD45.2 C57BL/6 Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox embryos and transplanted into lethally irradiated congenic CD45.1 C57BL/6 recipient mice. Four months after transplantation, recipient mice displayed decreased Mcm3Lox/Lox chimerism in all hematopoietic organs (Fig. 6d). Because formation of an adult HSC compartment was observed (Fig. 6d, right), donor-derived HSCs were re-isolated from the bone marrow of recipient mice to evaluate their proliferation potential in vitro and in vivo. Mcm3-deficient adult HSCs displayed RS markers γH2AX and RPA (Fig. 6e) and inefficient colony formation in methylcellulose in the presence of aphidicolin (Fig. 6f). Furthermore, when subjected to a major replication challenge such as a secondary transplantation, Mcm3Lox/Lox adult HSCs were drastically affected in their ability to self-renew and regenerate the blood system (Fig. 6g).
Discussion
Mcm2-7 are essential genes that encode key components of the main DNA helicase involved in genome replication. Mcm2-7 are highly expressed in proliferating cells, and DNA replication can still occur in cell lines after a significant reduction (>90% in some cases) in the cellular concentration of MCM protein complexes17,18,19. This tolerance to MCM downregulation is related to the fact that MCM complexes are engaged with DNA in large excess relative to the number of replication forks normally established during S phase. The surplus of MCM complexes license dormant origins that are activated only as a rescue mechanism when DNA replication is disrupted (reviewed in reference34). The use of dormant origins in vivo is well documented in the mouse, and MCM downregulation beyond ∼2/3 of its physiological levels causes embryonic lethality or promotes tumorigenesis in adults7,8,9,10,11,12,13. Therefore, it is likely that certain cell types in the developing embryo, such as stem and progenitor cells, are particularly sensitive to RS induced by low MCM levels.
We have tested the hypothesis that different cell types may have different requirements for MCM concentration using a novel strain with hypomorphic Mcm3 expression. While Mcm3Lox/Lox MEFs proliferated and replicated DNA with approximately 1/3 of the normal concentration of MCM3 protein, a similar reduction severely impaired hematopoietic progenitors, indicating a stricter requirement for origin licensing in the latter. In mid-gestation, hematopoiesis in the fetal liver is largely geared towards the production of RBCs to guarantee oxygen delivery to the rapidly growing embryo. Interestingly, erythroid precursors undergo several rounds of DNA replication and cell division during terminal differentiation, and genetic models that ablate cell cycle regulators such as Rb, E2F4, E2F8 or D-cyclins frequently result in embryonic anemia35,36,37,38. While D-Cyclin/CDK and Rb/E2F constitute the axis of a large transcriptional pathway regulating multiple genes, here we report for the first time that downregulation of a single MCM gene is sufficient to impair hematopoietic progenitor cells, causing anemia. Cytological analyses of embryonic blood revealed lower counts of RBCs and abundance of immature nucleated erythroblasts. Furthermore, transplantation of Mcm3-deficient fetal liver cells into lethally irradiated mice reconstituted the adult RBC population with much lower efficiency than Mcm3-competent cells. Our results indicate that a full complement of MCM is required for erythrocyte maturation in steady-state conditions and in response to erythropoietic stress. If MCM levels drop to approximately 1/3 of its normal concentration, the severity of the anemia causes embryonic lethality in the C57BL/6 genetic background. As reported in other mouse models such as the Mcm4-chaos mutant8,11,12 or the Rif1 KO mouse39, Mcm3Lox/Lox embryonic lethality was partially alleviated in a mixed C57BL/6-CD1 background and it was further rescued by overexpression of CHK1 kinase, reinforcing the connection between RS and the phenotypes observed.
To our knowledge, the single-molecule analyses of DNA replication in EB precursors isolated from the fetal liver provide the first evidence that the program of DNA replication undergoes active changes during the physiological maturation of mammalian erythrocytes. As pro-EBs proliferate and differentiate into mature reticulocytes, their replication program requires the progressive activation of more origins. This observation has interesting antecedents: chicken erythrocytic progenitors forced to differentiate ex vivo display a broadening in origin usage in the β-globin locus23. Also, origin firing is enhanced in murine erythroleukemia cells derived from transformed pro-EBs and forced to differentiate in culture40. It is conceivable that rapid DNA replication, driven from an increasingly high number of origins, is related to the rapid loss of DNA methylation marks observed during mouse erythropoiesis41. These patterns of DNA replication were altered in Mcm3-deficient pro-EBs, which displayed a higher frequency of origin activation and asymmetric forks from the start of their differentiation program. Later in differentiation, baso-EBs and chr-EBs were not capable of completing DNA replication, accumulated in S phase and activated the apoptotic program, thus preventing erythrocyte maturation. This likely triggers a compensation mechanism that pressures pro-EBs to replicate and proliferate, contributing to the aberrant replication patterns. Fork asymmetry reflects improper coordination between fork pairs42 and may also trigger the activation of compensatory origins. The fact that pro-EBs were capable of activating extra origins after Mcm3 downregulation underscores the fact that a very large excess of MCM proteins associate with DNA during G1.
Mid gestation is also the time of HSCs expansion in the fetal liver. At this stage, HSCs divide very rapidly (reviewed in reference43), which could also make them vulnerable to RS. The capacity of Mcm3Lox/Lox fetal HSCs to reconstitute the immune system upon transplantation into irradiated recipients was compromised but not completely impaired, and a population of donor-derived adult HSCs was generated in recipient mice. The lack of engraftment potential was strikingly exacerbated when Mcm3Lox/Lox HSCs were re-isolated from the bone marrow of recipient mice and tested in secondary transplantations. These observations are in line with the emerging concept that RS affects HSC functionality in older individuals. With age, HSCs lose reconstitution potential for causes that have been debated44,45,46,47,48 (reviewed in references49,50). Interestingly, a comparison of gene expression profiles of young versus old adult HSCs revealed that Mcm2-7 are downregulated in old HSCs, suggesting a new mechanistic link between faulty DNA replication and functional impairment32. The work presented here with a new mouse model with hypomorphic MCM expression provides direct genetic evidence that RS is a driving force behind HSC loss of functionality.
In summary, Mcm3 downregulation during embryonic development caused RS in hematopoietic progenitors leading to fetal anemia; in adults, it reduced life expectancy and promoted lymphomagenesis. Thus, future therapies designed to modulate RS are a promising way to fight ageing and hematopoietic malignancies.
Methods
Generation of Mcm3+/Lox mouse strain
A targeting vector was designed that included an IRES-EGFP-luciferase reporter cassette after the Mcm3 stop codon located at exon 17, two loxP sites flanking Mcm3 exons 14 to 17, and a FRT-flanked neomycin-resistance cassette for selection. The linearised vector was electroporated into 129Sv/Pas ES cells, and its genomic integration by recombination was screened by PCR and Southern blotting. Verified transgenic ES clones (Mcm3-Lox-Neo) were microinjected into C57BL/6J blastocysts and germline transmission was confirmed. Vector preparation and genetic manipulations were conducted at Genoway (Lyon, France). Mice carrying the Mcm3-Lox-Neo allele were crossbred with a strain expressing Flp recombinase to eliminate the neomycin-resistance cassette, generating the Mcm3-Lox allele. Mcm3+/Lox mice were crossbred with the CMV-Cre strain to generate the Mcm3-null allele, referred to as Mcm3−. Mice of both sexes were used in experiments, except when noted otherwise. Primers used for PCR genotyping are indicated in Supplementary Table 4.
Mouse handling and luciferase detection
Mice were hosted at the CNIO animal facility, except for a cohort of Mcm3+/Lox mice that were transferred to the UCSF animal facility for the analyses of HSCs. Animal procedures were approved by the Ethical Committee of the Instituto de Salud Carlos III (Madrid, Spain), and HSC transplantation experiments were performed under UCSF IACUC-approved protocols. For each experiment, sample size was calculated using Resource Equation51. In vivo luciferase imaging was performed in an IVIS imaging system (Caliper Life Sciences, CA) in mice anesthetized with 2.5% isoflurane, 10 min after an intraperitoneal injection of luciferin (150 mg kg−1 of body weight). Mice were partially shaved to monitor bioluminescence in the back skin. When indicated, mice were sacrificed 10 min after luciferin injection and necropsies were performed in order to monitor luciferase activity in internal organs.
Mouse embryonic fibroblasts (MEFs) isolation and culture
Mcm3+/+, Mcm3+/Lox and Mcm3Lox/Lox primary MEFs were derived from E12.5–14.5 embryos resulting from crosses between Mcm3+/Lox mice and cultured in Dulbecco’s modified minimal Eagle medium (DMEM) supplemented with 10% FBS and antibiotics. For cell proliferation curves, aliquots of 0.5 × 105 cells were seeded and counted every 2 days in a hemocytometer. When indicated, 10 μM bromo-deoxyuridine (BrdU; Sigma) was added to the medium for 30 min before cell harvesting. Cells were fixed in 70% ethanol, washed in PBS and stained with 50 μg ml−1 propidium iodide (Sigma) in the presence of 10 μg ml−1 RNase A (Qiagen). Fixed cells were treated with 2 M HCl for 20 min and incubated with FITC-conjugated anti-BrdU antibody for 60 min. Flow cytometry data was acquired in a FACS Canto II (BD, San Jose, CA) and analyzed with FlowJo 9.7.5 (Tree Star, Ashland, OR).
Quantitative RT–PCR, cell extracts and immunoblots
Total RNA was extracted using the RNeasy Mini Kit (Qiagen). 1 μg of total RNA was used for random-priming cDNA synthesis with SuperScript II (Invitrogen), and quantitative PCR was performed with Power SYBR Green master mix in an Applied Biosystems 7900HT Fast qRT–PCR machine. Primers used for gene expression are indicated in Supplementary Table 5. Extracts were prepared by direct suspension of cells in Laemmli buffer followed by three pulses of sonication for 15 s at 15% amplitude (Branson Digital Sonifier). SDS-PAGE and immunoblots were performed using standard methods.
Mcm3-Lox mRNA sequencing
Total RNA from Mcm3Lox/Lox MEFs was isolated and cDNA was prepared as indicated in the previous section. Three overlapping fragments covering the whole Mcm3-Lox cDNA molecule were amplified by PCR (primer sequences are shown in Supplementary Table 6) and sequenced. Experimental sequences were compared with the targeting vector used by Genoway and the NCBI reference sequence for Mcm3 mRNA (NM_008563.2). DNA and encoded protein alignments were performed with Clustal Omega (EMBL-EBI website).
Immunofluorescence microscopy
Primary MEFs or primary erythroid populations were incubated in μCLEAR bottom polylysine-treated 96-well or 384-well plates (Greiner Bio-One) for 2 h at 37 °C, fixed in 4% paraformaldehyde in PBS for 15 min at RT and permeabilizad with 0.5% Triton-X100 in PBS (5 min at RT). Cells were incubated in blocking solution (1% bovine serum albumin in PBS) for 30 min. Primary antibody solution was applied for 1 h at RT. For RPA immunostaining, soluble proteins were extracted prior to fixation with 0.5% Triton X-100 in CSK buffer (10 mM Pipes-KOH pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2). Cell nuclei were stained with DAPI (Sigma). Images were acquired either in a Leica-TCS SP5X confocal microscope, with a HCX PL APO 20x objective using LAS AF software or in an Opera High-Content Screening System (PerkinElmer) with an APO 20 ×, 0.7 NA water-immersion objective using Acapella software (PerkinElmer). To estimate the number of RPA foci-positive cells or γH2AX foci-positive cells, 100–200 or 250–500 cells were scored respectively in each condition. To measure intensity of MCM3 or γH2AX in erythroid populations, images were acquired from each well, nuclei were marked by DAPI staining and protein immunostaining intensity was measured within the nuclei.
Single-molecule analysis of DNA replication
Exponentially growing MEFs or single-cell suspensions of fetal livers were pulse-labeled with 50 μM CldU (20 min) followed by 250 μM IdU (20 min). Labeled cells were harvested and resuspended in 0.2 M Tris pH 7.4, 50 mM EDTA and 0.5% SDS. Stretched DNA fibers were prepared as described52. For immunodetection of labeled tracks, fibers were incubated with primary antibodies for 1 h at RT and the corresponding secondary antibodies for 30 min at RT, in a humidity chamber. DNA was stained with anti-ssDNA to assess fiber integrity. Fiber images were obtained in a DM6000 B Leica microscope with an HCX PL APO 40 ×, 0.75 NA objective. The conversion factor used was 1 μm=2.59 kb. In each assay, >300 individual tracks were measured for FR estimation, >100 fibers containing two or more origins were analyzed for IOD estimation, >200 labeled fibers were counted for fiber length and origin density, and >180 bidirectional forks were counted for fork symmetry.
Histology and immunohistochemistry (IHC)
Embryos, normal tissues and tumour samples were fixed in 10% buffered formalin (Sigma) and embedded in paraffin using standard procedures. For histopathological studies, 3 μm sections were stained with haematoxylin and eosin (HE). The following antibodies were used for IHC analysis: MCM3 (generated in rabbits immunized with synthetic peptide N-CSQEDTEQKRKRRK-C conjugated to KLH; Sigma-Genosys, UK; serum was used at 1:500 dilution), Ki67 (Master Diagnostica, 0003110QD; 1:500), γH2AX (Millipore, 05-636; 1:100), pS15-p53 (Cell Signaling, 9284; 1:200), Ter119 (BD, 550565; 1:50), Pax5 (Santa Cruz Biotechnology, sc-1974; 1:50), CD3 (Santa Cruz Biotechnology, sc-1127; 1:50) and FVIII (Dako, A0082; 1:200). Tissue slides were digitalized using a Mirax scanner (Carl Zeiss) and equivalent areas per tissue and group were analyzed using AxioVision digital image processing software (Carl Zeiss). Areas of positive staining were normalized to the total analyzed area.
Fluorescence activated cell sorting of hematopoietic cells
To prepare fetal liver cell suspensions, E14.5–16.5 fetal livers were disaggregated in 2% FBS-RPMI by multiple passages through a 26-gauge needle and filtered through a 70 μm strainer (BD Falcon). c-kit-positive cells were enriched by positive selection using streptavidin-conjugated magnetic beads and an autoMACS cell separator (Miltenyi Biotec, Auburn, CA). Depletion of Lineage (Lin)+ cells was done by incubation with purified Lin antibodies (see below) followed by magnetic beads (Dynal, Life Technologies) as per manufacturer instructions. HSCs were stained with unconjugated Lin antibodies Gr-1 (1:800), Mac1 (1:100), B220 (1:800), CD3 (1:100), CD4 (1:100), CD5 (1:100), CD8 (1:100), Ter-119 (1:100), obtained from the UCSF Hybridoma Core Facility), goat anti-rat-PE-Cy5 (Invitrogen, A10691; 1:400), c-Kit-APC-eFluor780 (eBioscience, 47-1171-82; 1:800), Sca-1-PB (BioLegend, 108120; 1:400), Flk2-Bio (eBioscience, 13-1351-82; 1:100), CD48-Alexa Fluor 647 (BioLegend, 103416; 1:400), CD150-PE (BioLegend, 115904; 1:400) and SA-PE-Cy7 (eBioscience, 25-4317-82; 1:800) antibodies. FcγR-PerCP-eFluor710 (eBioscience, 46-0161-82; 1:400) and CD34-FITC (eBioscience, 11-0341-85; 1:25) were included for concomitant isolation of MPPs and GMPs, and granulocytes were stained with Mac-1-PE-Cy7 (eBioscience, 25-0112-82; 1:3200) and Gr-1-PB (eBioscience, 57-5931-82; 1:400) antibodies. Stained cells were re-suspended for final analysis in Hanks Buffered Salt Solution (HBSS) with 2% heat-inactivated fetal calf serum (FCS) and 1 μg/ml propidium iodide (PI) for dead cell exclusion. Cell isolation was performed on a FACSAriaII (BD, San Jose, CA) and cell analysis was performed on a LSRII (BD, San Jose, CA).
E13.5-15.5 erythroid populations were characterized using Lineage-APC (1:100), Ter119-FITC (1:200) and CD71-PE (1:200) conjugated antibodies from Pharmingen (San Jose, CA). Stained cells were resuspended in PBS, 0.2% BSA, 3 mM EDTA and run in a LSR Fortessa (BD, San Jose, CA). Pulse processing was used to exclude cell aggregates and 1 μg/ml DAPI was added for dead cell exclusion. At least 20,000 events were collected from the single alive population. For DNA content analyses, fetal liver suspensions were fixed in 4% paraformaldehyde in PBS for 15 min at RT. After fixation, cells were washed in PBS and stained with 5 μg/ml Hoescht (Invitrogen). For apoptosis assays, cells were labelled with surface markers Ter119 and CD71, fixed, permeabilized and labelled with anti-activated Caspase 3 (BD Biosciences; 1:200). Cell sorting was performed on a FACSAriaII (BD, San Jose, CA) and data was analyzed with FlowJo 9.7.5 (Tree star, Ashland, OR).
Blood sampling
After embryo exsanguination, peripheral blood was washed and analyzed for erythrocyte parameters on an Abacus Junior Vet Hematology Blood Analyzer (Diatron) or deposited onto coverslips using a Thermo-Fisher Shandon Cytospin 2 (600 r.p.m. 5 min, mounted on slides and stained with Giemsa as described28. To determine the percentage of nucleated cells in E14.5 embryonic blood samples, cells were stained with Ter119-FITC (1:200; Pharmingen, San Jose, CA) and nucleated/enucleated cells were differentiated with 5 μg ml−1 Hoescht (Invitrogen).
Steady-state and stress erythropoiesis response
Mice with constitutive expression of fluorescent tdTomato from the Rosa26 locus were generated by germline recombination of the Rosa-CAG-LSL-tdTomato allele29, carried by the Ai14 reporter mouse line (stock 007914, Jackson Laboratory). Total cells were recovered from the bone marrow of CAG-Tomato mice and from the fetal livers of E14.5 Mcm3+/+ or Mcm3Lox/Lox C57Bl/6 donor mice, and transplanted into lethally irradiated C57BL/6 primary female recipients (106 cells per mouse; 5 mice per group). 8 weeks post-transplantation, donor chimerism (Tom+/Tom-) was analyzed in peripheral blood after staining with Ter119. To monitor the erythropoietic stress response, mice were subjected to phleblotomy (400 μl peripheral blood per 25 g of body weight). 4 days post-phlebotomy, accumulation of RBC progenitors EryA, EryB and EryC in the spleen was monitored as described30, differentiating between Tom+ and Tom- cells.
HSC transplantation
Fetal HSCs (Lin-/Sca-1+/c-Kit+/Flk2-/CD150+/CD48-) were isolated from the fetal livers of E14.5 Mcm3+/+, Mcm3+/Lox or Mcm3Lox/Lox CD45.2 C57Bl/6 donor mice, and transplanted into lethally irradiated CD45.1 C57Bl/6 primary recipients (50 fetal HSCs per mouse; 5 mice per group) together with 300,000 Sca-1-depleted CD45.1 helper BM cells. 4 months post-transplantation, the primary recipients were sacrificed to assess the percentage of donor-derived chimerism in the bone marrow (BM), spleen, peripheral blood and HSC compartment and to re-isolate donor-derived adult MCM3+/+ or MCM3Lox/Lox HSCs53. Donor chimerism was analyzed using CD45.2-FITC, (eBioscience 12-0453-83; 1:50), B220-APC-e780 (47-0452-82; 1:800), Gr-1-PB (1:400), Mac-1-PE-Cy7 (1:3200), CD3-e660 (eBioscience, 50-0032-82; 1:400) and Ter-119-PE-Cy5 (eBioscience 15-5921-83; 1:800) antibodies. Re-isolated CD45.2+ donor-derived adult MCM3+/+ or MCM3Lox/Lox HSCs were transplanted into lethally irradiated CD45.1 secondary recipients (500 HSCs per mouse; 5 mice per group) together with 300,000 Sca-1-depleted CD45.1 helper BM cells.
Single-cell tracking of division kinetics
HSCs isolated from the fetal liver of E14.5–16.5 embryos were sorted into 96-well plates (1 cell/ well) and visually inspected after 12 h to confirm successful single cell sort. At the indicated time points, cell cultures were monitored to establish the kinetics of the first cell division (appearance of 2 or more cells). 96 wells were scored per condition.
HSC colony formation
100 HSCs were cultured in Iscove’s modified Dulbecco’s media (IMDM) supplemented with 5% FBS (StemCell Technology, 06200), 1 × penicillin/streptomycin, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 2 mM L-glutamine and 50 μM 2-mercaptoethanol (base media) supplemented with the following cytokines (all from PeproTech): IL-3 (10 ng ml−1), GM-CSF (10 ng ml−1), SCF (25 ng ml−1), IL-11 (25 ng ml−1), Flt-3L (25 ng ml−1), Tpo (25 ng ml−1) and Epo (4 U ml−1) (culture media). When indicated, 50 ng ml−1 aphidicolin (Sigma, A4487) was added to the culture media. Colonies (triplicates) were counted on day 7. All cultures were maintained at 37 °C in a 5% CO2 water jacket incubator (Thermo Scientific).
Statistical methods
In column graphs, data are expressed as mean±s.d. Statistical analyses were done using Fisherŕs test. When the data are presented in scatter dot plots, the bar corresponds to the median value. For the analyses of FR, IOD and fork symmetry parameters in stretched DNA fibers and immunofluorescence data from acquired in the high-throughput Opera system, data distribution was normally not Gaussian, and differences between samples were assessed with the nonparametric Mann-Whitney rank-sum test. In the analysis of apoptosis and cell death in EB populations, the mean values derived from 3 independent experiments were compared by One-Way Anova and further Bonferroni post-test. Statistical analysis was performed in Prism v4.0 (GraphPad Software).
Additional information
How to cite this article: Alvarez, S. et al. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat. Commun. 6:8548 doi: 10.1038/ncomms9548 (2015).
References
Costa, A., Hood, I. V. & Berger, J. M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 82, 25–54 (2013).
Masai, H., Matsumoto, S., You, Z., Yoshizawa-Sugata, N. & Oda, M. Eukaryotic chromosome DNA replication: where, when and how? Annu. Rev. Biochem. 79, 89–130 (2010).
Bicknell, L. S. et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 43, 350–355 (2011a).
Bicknell, L. S. et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 43, 356–359 (2011b).
Guernsey, D. L. et al. Mutations in origin recognition complex gene ORC4 cause Meier–Gorlin syndrome. Nat. Genet. 43, 360–364 (2011).
Gineau, L. et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency and natural killer cell deficiency. J. Clin. Invest. 122, 821–832 (2012).
Pruitt, S. C., Bailey, K. J. & Freeland, A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells 25, 3121–3132 (2007).
Shima, N. et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 39, 93–98 (2007).
Chuang, C. H., Wallace, M. D., Abratte, C., Southard, T. & Schimenti, J. C. Incremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet. 6, e1001110 (2010).
Kunnev, D. et al. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene 29, 3630–3638 (2010).
Kawabata, T. et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell. 41, 543–553 (2011a).
Kawabata, T. et al. A reduction of licensed origins reveals strain-specific replication dynamics in mice. Mamm. Genome 22, 506–517 (2011b).
Bagley, B. N. et al. A dominantly acting murine allele of Mcm4 causes chromosomal abnormalities and promotes tumorigenesis. PLoS Genet. 8, e1003034 (2012).
Williams, G. H. & Stoeber, K. The cell cycle and cancer. J. Pathol. 226, 352–364 (2012).
Tsaniras, S. C. et al. Licensing of DNA replication, cancer, pluripotency and differentiation: an interlinked world? Semin. Cell. Dev. Biol. 30, 174–180 (2014).
Blow, J. J., Ge, X. Q. & Jackson, D. A. How dormant origins promote complete genome replication. Trends. Biochem. Sci. 36, 405–414 (2011).
Woodward, A. M. et al. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 173, 673–683 (2006).
Ge, X. Q., Jackson, D. A. & Blow, J. J. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21, 3331–3341 (2007).
Ibarra, A., Schwob, E. & Méndez, J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl Acad Sci. USA 105, 8956–8961 (2008).
Hyrien, O., Maric, C. & Mechali, M. Transition in specification of embryonic metazoan DNA replication origins. Science 270, 994–997 (1995).
Sasaki, T., Sawado, T., Yamaguchi, M. & Shinomiya, T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19, 547–555 (1999).
Gregoire, D., Brodolin, K. & Mechali, M. HoxB domain induction silences replication origins within the locus and specifies a single origin at its boundary. EMBO. Rep. 7, 812–816 (2006).
Dazy, S., Gandrillon, O., Hyrien, O. & Prioleau, M. N. Broadening of DNA replication origin usage during metazoan cell differentiation. EMBO Rep. 7, 806–811 (2006).
Norio, P. et al. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell. 20, 575–587 (2005).
Mechali, M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell. Biol. 11, 728–738 (2010).
Norio, P. DNA replication: the unbearable lightness of origins. EMBO Rep. 7, 779–781 (2006).
Pop, R. et al. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 21, 8 (2010).
Fraser, S. T., Isern, J. & Baron, M. H. Maturation and enucleation of primitive erythroblasts during mouse embryogenesis is accompanied by changes in cell-surface antigen expression. Blood 109, 343–352 (2007).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Liu, Y. et al. Suppression of Fas-FasL coexpression by erythropoietin mediates erythroblast expansion during the erythropoietic stress response in vivo. Blood 108, 123–133 (2006).
Lopez-Contreras, A. J., Gutierrez-Martinez, P., Specks, J., Rodrigo-Perez, S. & Fernandez-Capetillo, O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J. Exp. Med. 209, 455–461 (2012).
Flach, J. et al. Replication Stress is a Potent Driver of Functional Decline in Aging Hematopoietic Stem Cells. Nature 512, 198–202 (2014).
Wang, J. et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148, 1001–1014 (2012).
Alver, R. C., Chadha, G. S. & Blow, J. J. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA. Repair. (Amst). 19, 182–189 (2014).
Kozar, K. et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118, 477–491 (2004).
Kinross, K. M., Clark, A. J., Iazzolino, R. M. & Humbert, P. O. E2f4 regulates fetal erythropoiesis through the promotion of cellular proliferation. Blood 108, 886–895 (2006).
Sankaran, V. G., Orkin, S. H. & Walkley, C. R. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 22, 463–475 (2008).
Hu, T. et al. Concomitant inactivation of Rb and E2f8 in hematopoietic stem cells synergizes to induce severe anemia. Blood 119, 4532–4542 (2012).
Chapman, J. R. et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 (2013).
Monturus, E. et al. DNA replication fading as proliferating cells advance in their commitment to terminal differentiation. Sci. Rep. 2, 279 (2012).
Shearstone, J. R. et al. Global DNA demethylation during mouse erythropoiesis in vivo. Science 334, 799–802 (2011).
Conti, C. et al. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell 18, 3059–3067 (2007).
Pietras, E. M., Warr, M. R. & Passegué, E. Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720 (2011).
Rossi, D. J. et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad Sci. USA 102, 9194–9199 (2005).
Rossi, D. J. et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007).
Chambers, S. M. et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5, e201 (2007).
Mohrin, M. et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185 (2010).
Beerman, I., Seita, J., Inlay, M. A., Weissman, I. L. & Rossi, D. J. Quiescent Hematopoietic Stem Cells Accumulate DNA Damage during Aging that Is Repaired upon Entry into Cell Cycle. Cell Stem Cell 15, 37–50 (2014).
Rossi, D. J., Jamieson, C. H. & Weissman, I. L. Stems cells and the pathways to aging and cancer. Cell 132, 681–696 (2008).
Geiger, H., de Haan, G. & Florian, M. C. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 13, 376–389 (2013).
Festing, M. F. Design and statistical methods in studies using animal models of development. ILAR. J. 47, 5–14 (2006).
Mourón, S. et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389 (2013).
Warr, M. R. et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327 (2013).
Acknowledgements
We thank members of our laboratories for helpful discussions, Marcos Malumbres (CNIO) for advice on the design of the Mcm3-Lox allele, Isabel Blanco for her administrative help with mouse work and Soraya Ruiz for excellent handling of the mouse colony in J.M.’s group. Research was supported by the Spanish Ministry of Economy and Competitiveness (grants BFU2013-49153-P and Consolider-Ingenio CSD2007-00015 to J.M., SAF2011-23753 to O.F.-C., BFU2012-35892 to J.I.) and RO1 HL092471 to E.P., S.A. was the recipient of an EMBO short-term fellowship to visit E.P.’s laboratory at UCSF. We are grateful to Manuel Serrano (CNIO), Almudena Ramiro (CNIC) and Arkaitz Ibarra (The Salk Institute, USA) for useful comments on the manuscript.
Author information
Authors and Affiliations
Contributions
S.A., M.D. and J.F. performed most of the experiments; S.R.A. confirmed the integrity of the MCM3-Lox allele and contributed to the DNA fiber experiments; A.J.L.C. and O.F-.C. provided the super-Chk1 strain; D.M. contributed to all flow cytometry analyses; M.C. performed pathology analyses; J.I. provided Tom+ mice and contributed to the erythropoiesis experiments; J.F. and E.P. characterized the functionality of Mcm3-deficient HSCs; JM designed the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-9 and Supplementary Tables 1-6 (PDF 1120 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Alvarez, S., Díaz, M., Flach, J. et al. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat Commun 6, 8548 (2015). https://doi.org/10.1038/ncomms9548
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms9548
This article is cited by
-
The N6-methyladenosine methyltransferase METTL16 enables erythropoiesis through safeguarding genome integrity
Nature Communications (2022)
-
Efficient terminal erythroid differentiation requires the APC/C cofactor Cdh1 to limit replicative stress in erythroblasts
Scientific Reports (2022)
-
Dynamics of replication origin over-activation
Nature Communications (2021)
-
Pediatric MDS and bone marrow failure-associated germline mutations in SAMD9 and SAMD9L impair multiple pathways in primary hematopoietic cells
Leukemia (2021)
-
Fanconi anemia proteins participate in a break-induced-replication-like pathway to counter replication stress
Nature Structural & Molecular Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.