Abstract
The genetic basis of sporadic colorectal cancer (CRC) is not well explained by known risk polymorphisms. Here we perform a meta-analysis of two genome-wide association studies in 2,627 cases and 3,797 controls of Japanese ancestry and 1,894 cases and 4,703 controls of African ancestry, to identify genetic variants that contribute to CRC susceptibility. We replicate genome-wide statistically significant associations (P<5 × 10−8) in 16,823 cases and 18,211 controls of European ancestry. This study reveals a new pan-ethnic CRC risk locus at 10q25 (rs12241008, intronic to VTI1A; P=1.4 × 10−9), providing additional insight into the aetiology of CRC and highlighting the value of association mapping in diverse populations.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer deaths in the United States. Genetics is known to play an important role in CRC susceptibility1. However, genome-wide association studies (GWASs), mostly conducted in European-descent populations, have only identified 30 common risk variants (22 independent loci) for CRC, markedly fewer than for prostate or breast cancer.
To discover additional risk loci for this cancer, we combine, via a meta-analysis, two GWASs of CRC in populations of Japanese and African American ancestry. The top associations for single-nucleotide polymorphisms (SNPs) in VTI1A are replicated in European-descent populations.
Results
In the first GWAS, Japanese samples (n=6,424) were identified from the Multiethnic Cohort study (MEC), the Colorectal Cancer Family Registry (CCFR), the Japan Public Health Center cohort study (JPHC) and three case–control studies in Hawaii (CR2&3) and in Fukuoka and Nagano, Japan (Supplementary Table 1). Blood leukocyte DNA samples were genotyped on the Illumina 1M-Duo or the Illumina 660W-Quad arrays, yielding, after quality control (QC) procedures, data for 323,852 SNPs available for all Japanese samples (see Methods and Supplementary Methods). Un-typed markers or markers with partly missing values were imputed with BEAGLE2 using East Asians from the 1000 Genomes Project (phase 1, release 3) as the reference panel. The second GWAS of African American samples (n=6,597) (Supplementary Table 2) were identified from the MEC, CCFR, the Southern Community Cohort Study (SCCS), the MD Anderson Cancer Center, the University of North Carolina CanCORS study (UNC-CanCORS) and Rectal Cancer Study (UNC-Rectal), and from the Prostate, Lung, Colorectal and Ovarian Cancer Screening (PLCO) Trial. African American samples were genotyped using the Illumina 1M-Duo bead arrays (except 170 PLCO subjects on Illumina Omni 2.5M). Imputation was performed with BEAGLE using Europeans and Africans from the 1000 Genomes Project (phase 1, release 3) as reference panels. Over 4.2 million genotyped or imputed autosomal markers were available for both studies.
In both GWASs, cases and controls were well matched with regard to genetic ancestry based on principal component (PC) analyses (Methods and Supplementary Figs 1 and 2). We used logistic regression within each ethnic group to test for the association of SNP dosage with CRC risk, adjusting for age at blood draw, sex and the first four PCs. The genomic control3 inflation factor (λ) was 1.04 for each individual study, indicating little effect of population stratification after controlling for global ancestries (Supplementary Fig. 3).
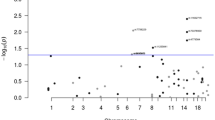
After combining the two GWASs, we observed three SNPs in the VTI1A gene on chromosome 10q25 to be statistically significant at the genome-wide significance level (P<5 × 10−8; Fig. 1; Table 1). The strongest association was for rs12241008 (114,280,702 bp) (odds ratio (OR)=1.19, 95% confidence interval (CI) 1.12–1.26, P=2.9 × 10−8, allele frequency 0.19 and 0.25 in African Americans and Japanese, respectively), with highly consistent associations in both populations (I2=0). The other two SNPs, rs7894915 (114,277,039 bp, P=4.8 × 10−8) and rs10082356 (114,278,181 bp, P=4.9 × 10−8), were in high linkage disequilibrium (LD) with rs12241008 (r2 from 0.80 to 1.0 in East Asians, Africans and Europeans), with risk estimates almost identical to those for rs12241008 (Supplementary Tables 3 and 4). This locus has not previously been reported to be associated with CRC.
Results in the Japanese (a) (n=6,424), African Americans (b) (n=6,595) and in the combined data (Japanese and African Americans) (c) are displayed. The SNP with the smallest P-value from meta-analysis in the combined data (n=13,019), rs12241008, is shown as a purple diamond. r2 is in relation to this SNP from the 1000 Genomes Project in East Asians (a,c) or in Africans (b). The plots were generated using LocusZoom26.
We subsequently replicated these associations in two large CRC consortia of European-descent populations (allele frequency 0.09): Colorectal Transdisciplinary Study (CORECT) with 7,561 cases and 6,328 controls from eight participating studies (combined OR=1.09, P=0.036, Table 1), and the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) with 9,262 cases and 11,883 controls from 18 participating studies (combined OR=1.09, P=0.018, Table 1). The combined P-value for rs12241008 in the Japanese, African Americans and Europeans was 1.4 × 10−9 (OR=1.13, 95% CI 1.09–1.18). A meta-analysis using individual study-level statistics yielded similar results (P=1.5 × 10−9). Although risk estimates were consistent across individual studies (I2=8%, Phet=0.35, d.f.=27) (see the forest plot in Supplementary Fig. 4), there was some evidence for heterogeneity in effects across ethnic groups (I2=53%, Phet=0.12, d.f.=2). This possible heterogeneity in effects, along with the low allele frequencies observed in European-descent populations (and therefore low power), could partially explain why previous GWASs in Europeans failed to identify this locus, and thus emphasizes the importance of conducting GWASs in ethnically diverse populations.
Similar results were observed for rs7894915 and rs10082356 when the data were combined with the European-descent GWASs (P=1.6 × 10−9 and 1.5 × 10−9, respectively). Nine other SNPs in this region (located within 12 kb in the same LD block as rs12241008 in East Asians, Africans and Europeans, Supplementary Fig. 5) also had P-values<5 × 10−8 when all data were combined (Supplementary Tables 3 and 4). However, the strongest association signal was still with rs12241008 (Supplementary Fig. 5) and none of the other nearby SNPs within 200 kb represented an independent signal after conditioning on rs12241008 in the African American and Japanese GWASs. Results for these 12 SNPs were similar (change in OR <1.2%) with or without adjustment for local ancestry estimates among African Americans (Methods).
There was no important heterogeneity in ORs in the Japanese or the African American data by anatomical site (colon versus rectal cancer) (P-values>0.74), by stage (regional/distant versus local/in situ) (P-values>0.6), by age of diagnosis (P-values>0.7) or by sex (P=0.04 in African Americans and 0.80 in Japanese) for the most significant marker rs12241008 (stratified analysis results are in Supplementary Table 5).
No association reached the genome-wide significance threshold (5 × 10−8) in the Japanese GWAS when analysed separately. One SNP on chromosome 7, rs79453636, passed this threshold (P=2.9 × 10−8) in the African American study (Supplementary Fig. 3). However, this association was not replicated (Supplementary Table 6) in the Japanese or in the combined European-descent data (P-values>0.15).
Out of 30 known CRC susceptibility SNPs, 27 were available for analysis in the Japanese study and 23 (85%) effects were in the same direction as in the original GWAS reports (Supplementary Table 7). Replication and fine-mapping of the known risk loci in the African American study was summarized previously4. Twelve out of the 27 associations were replicated in the meta-analysis of the Japanese and African American data (P<0.05) and 23 risk estimates were directionally consistent with those originally reported (Supplementary Table 7). Considerable heterogeneity in disease risk between the two ethnic groups (I2>50%) was observed for six SNPs (Supplementary Table 7).
The three genetic variants with the strongest association with CRC, rs12241008, rs7894915 and rs10082356, are located in intron 3 of the VTI1A gene, which encodes vesicle transport through interaction with t-SNAREs 1A. VTI1A is involved in regulating insulin-stimulated trafficking of secretory vesicles enriched with both GLUT4 (glucose transporter) and Acrp30 in adipocytes5; it also plays key roles in neuronal development6 and in selectively maintaining spontaneous neurotransmitter release7. A recent GWAS in never-smoking Asian women has identified rs7086803 in intron 7 of VTI1A as a lung cancer susceptibility variant (218 kb from and not in LD with rs12241008)8. Interestingly, a gene fusion product, VTI1A-TCF7L2, was identified in colorectal tumours and shown to promote anchorage-independent growth of cultured tumour cells9. The fusion occurs between VTI1A exon 3 (chr10: 114,220,869) and TCF7L2 exon 4 (chr10: 114,760,545) and results in the deletion of both the intron 3 CRC and intron 7 lung cancer risk variants. No coding variant is in LD with the top three SNPs. Among the three VTI1A SNPs associated with CRC in this study, rs7894915 and rs10082356 lie in predicted transcriptional regulatory regions, suggesting enhancer and promoter regulatory activities across multiple cell lines (Supplementary Table 8)10. We explored regulatory effects of the SNPs correlated with rs12241008 in a cis-expression quantitative trait loci analysis in 40 paired colon adjacent-normal and tumour tissue samples from European descent patients11. Among the SNPs in high LD (r2>0.8) with rs12241008 in East Asians, the intronic SNP rs7081965 (alleles: A/T) affected VTI1A expression (P=0.003) in colon tumour tissue. Rs7081965 is also in considerable LD with rs12241008 in Africans (r2=0.21, |D′|=0.88) and in Europeans (r2=0.24, |D′|=1) in the 1000 Genomes data. Although the association of rs7081965 with CRC was not statistically significant in this study (OR=1.09 for allele T, P=8.2 × 10−6 from the three ethnic groups combined), these results provide an interesting lead for future functional investigations.
In summary, this trans-ethnic GWAS identified a new CRC susceptibility locus at 10q25 with directionally consistent associations across three ethnic/racial populations, providing additional insight into the genetic architecture of CRC. Further work is needed to dissect this genetic signal and to conduct functional studies to uncover the mechanisms underlying this association.
Methods
Japanese subjects and QC on genotypes
Details on study design and basic characteristics for each study are provided in Supplementary Methods. Briefly, 1,703 MEC Japanese American subjects were genotyped by the Broad Genotyping Center on the Illumina 1M-Duo Array and 1,602 (803 cases, 799 controls) passed their initial QC filters. To maximize sample size, initially ‘failed’ samples on five plates were re-clustered with a customized genotype calling algorithm—this step recovered 42 additional MEC subjects (23 cases, 19 controls), although not all SNPs on the array were preserved. To increase statistical power and to provide a larger control pool, 1,033 prostate cancer-free men and 808 breast cancer-free women genotyped on the Illumina 660W‐Quad platform were drawn from the MEC prostate cancer12 and breast cancer13 studies, respectively.
Japanese from the following studies were all genotyped on the Illumina 1M-Duo array by the University of Southern California (USC) Epigenome Center: 697 from CCFR (384 cases, 313 controls), 155 cases from CR2&3, 1,463 from Fukuoka, Japan (685 cases, 778 controls), 212 from Nagano, Japan (106 cases, 106 controls) and 1,332 from JPHC (670 cases, 662 controls). In general, all genotyped samples were examined and excluded according to the following: (1) call rates<90%, 95% or 97% depending on the batches, (2) missing on basic covariates (age, sex or disease status), (3) gender mismatch, that is, the reported sex was different from that estimated based on X chromosome inbreeding coefficient F, calculated by PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/), (4) ethnicity outliers, that is, subjects fell out of the Japanese cluster (by visual inspection) on PC plots, where PCs were derived for study subjects as well as unrelated HapMap CEU, YRI and JPT samples with our own R program (The Comprehensive R Archive Network http://www.r-project.org/), based on about 20k SNPs with inter-marker distance>100 kb, and (5) close (⩾2nd degree) relatives, where relationships were derived from estimated probabilities of sharing 0, 1 or 2 alleles based on genomic data (calculated by PLINK), and relatives were removed in the following order: subjects with most relatives, controls and subjects with lower call rates. All cases were verified by histological records to have invasive carcinoma of the colon or rectum. More details on genotype QC can be found in Supplementary Methods. After QC, the following subjects were retained in analysis: 3,094 from the MEC (797 cases, 2,297 controls), 285 from CCFR (276 cases and 9 controls), 134 cases from CR2&3, 1,411 from Fukuoka, Japan (662 cases, 749 controls), 207 from Nagano, Japan (105 cases, 102 controls) and 1,293 from the JPHC (653 cases, 640 controls).
African American subjects and QC on genotypes
Sample collection and genotyping QC have been described in detail elsewhere4 and in Supplementary Methods. We genotyped 7,168 African American samples from six studies/centres: the MEC (442 cases, 4,620 controls), CCFR (999 cases, 290 controls), SCCS (164 cases, 160 controls), the MD Anderson Cancer Center (189 cases), UNC-CanCORS (84 AA cases) and UNC-Rectal (112 cases, 108 controls) on the Illumina 1M-duo platform. QC procedures for all subjects were similar to the criteria described for the Japanese study subjects. Included in analysis were 6,427 subjects (4,609 controls, 1,818 cases) on 1,049,327 markers. We also included 170 PLCO samples (76 cases, 94 controls) that were previously genotyped on the Illumina Omni 2.5M array and pre-filtered by the NCI genotyping centre for analysis (527,383 markers that overlapped with other studies). Overall, 6,597 subjects (1,894 cases, 4,703 controls) were used in association testing. Supplementary Table 2 shows the distribution of subjects by participating study.
Imputation
Prediction of un-typed or partly genotyped SNPs was performed with BEAGLE 3.3 (ref. 2) using the 1000 Genomes Project (phase 1, release 3) East Asians as reference panels for the Japanese data and Europeans and Africans for the African American data. Imputation was performed separately for the two ethnic groups with all cases and controls combined. Markers with minor allele frequencies<0.005 in reference panels were excluded from imputation. For the African American data, 10,050,748 markers with imputation accuracy R2>0.8 were kept for association analysis; for Japanese data, 4,266,108 markers with imputation R2>0.95 were retained. Altogether, 4,276,079 autosomal genotyped or imputed markers were available in both populations for meta-analysis.
Analysis of the Japanese and African American GWASs
PCs were calculated as in EIGENSTRAT14 with our own R program, including unrelated HapMap CEU, YRI and JPT samples as population controls. Ethnicity outliers were identified on PC plots by visual inspection and subsequently removed. Pair-wise PC plots suggested that the first two PCs were most informative for global ancestry and the distribution of PCs was similar among all cases and controls in both Japanese and African Americans (Supplementary Figs 1 and 2). Logistic regression of CRC on allelic dosage with adjustment for age at blood draw, sex and the first four PCs was performed to obtain OR estimates and 95% CI of per increase in allele count with PLINK, where age was grouped as <55 years, 5-year intervals from 55 to 80 and ⩾80 years. The genomic control factor (λ) was estimated from the median of the χ2 statistics divided by 0.456.
Heterogeneity of genetic effects by site (colon versus rectal cancer, mutually exclusive), stage (regional/distant versus local/in situ) and age at diagnosis (≤55 versus >55 years) was tested in a case-only analysis. Effect modification by sex was assessed comparing the model with and without the cross-product term. These and additional stratified analyses by site, stage, age at diagnosis and sex were adjusted for age at blood draw, sex (where appropriate), the first four PCs and BMI.
Conditional analyses were performed to examine the independence of association signals in the chromosome 10 region, conditioning on the SNP with the smallest P-value. Significance of the additional contribution by other SNPs was calculated based on a likelihood ratio test. These analyses were carried out using SAS 9.3.
Local ancestry estimation for African Americans
The percentage of African ancestry (0, 50 or 100%, that is, half of the estimated number of African chromosomes) was inferred for each participant at the putative CRC risk locus on chromosome 10 (±250 kb) with the LAMP program v2.4 (ref. 15). To summarize local ancestry, for each individual we averaged across all local ancestry estimates that are within the region. The effect of local ancestry was evaluated by examining the relative change in ORs with and without adjustment for local ancestry in logistic regression.
CORECT study for replication
The CORECT study meta-analysis was conducted using germline DNA in the Molecular Epidemiology of Colorectal Cancer study (MECC) (set 1: 484 cases and 498 controls; set 2: 1,120 cases and 820 controls), CCFR (set 1: 1,977 cases and 999 controls; set 2: 1,660 cases and 1,393 controls), Kentucky case–control study (1,038 cases and 1,134 controls), Newfoundland case–control study (548 cases and 538 controls), American Cancer Society CPS II nested case–control study (ACS/CPSII, 539 cases and 469 controls) and the Melbourne nested case–control study (195 cases and 477 controls). All subjects were self-reported whites. The majority of the studies were genotyped using the Affymetrix Axiom CORECT Set containing ~1.3 million SNPs and indels on two physical genotyping chips (Supplementary Table 3). Genotype data were screened based on filters such as call rates, concordance rates, sample relatedness and ethnic outliers. IMPUTE2 (ref. 16) was used to impute missing genotypes based on the cosmopolitan panel of reference haplotypes from Phase I of the 1000 Genomes Project. Imputed genotypes were screened based on stringent imputation quality and accuracy filters (info⩾0.7, certainty⩾0.9, concordance⩾0.9 between directly measured and imputed genotypes after masking input genotypes for genotyped markers only). Associations between genetic variants and CRC risk were tested using a log-additive genetic model within each study, allowing for study-specific adjustment for age, sex, study centre, genotyping batch and 2–4 PCs. More details of each participating study can be found in Supplementary Methods.
GECCO study for replication
The GECCO GWAS consortium has been described before17,18,19. The consortium consisted of European-descent participants within the French Association Study Evaluating RISK for sporadic CRC (ASTERISK, 948 cases and 947 controls); CR2&3 (87 cases and 125 controls); Darmkrebs: Chancen der Verhütung durch Screening (DACHS set 1: 1,710 cases and 1,707 controls; DACHS set 2: 675 cases and 498 controls); Diet, Activity, and Lifestyle Study (DALS set 1: 706 cases and 710 controls; DALS set 2: 410 cases and 464 controls); Health Professionals Follow-up Study (HPFS set 1: 227 cases and 230 controls; HPFS set 2: 176 cases and 172 controls); MEC (328 cases and 346 controls); Nurses’ Health Study (NHS set 1: 394 cases and 774 controls; NHS set 2: 159 cases and 181 controls); Ontario Familial Colorectal Cancer Registry (OFCCR, 650 cases and 522 controls); Physician’s Health Study (PHS, 382 cases and 389 controls); Postmenopausal Hormone study (PMH, 280 cases and 122 controls); Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO set 1: 533 cases and 1,976 controls; PLCO set 2: 486 cases and 415 controls); VITamins And Lifestyle (VITAL, 285 cases and 288 controls); and the Women’s Health Initiative (WHI set 1: 470 cases and 1,529 controls; WHI set 2: 1,006 cases and 1,010 controls). All individual studies were genotyped on Illumina arrays on 240k–730k markers and went through rigorous QC. The genotype data were imputed to increase the density of genetic variants. The haplotypes from the 1000 Genomes Project Phase I were used as the reference panel. Logistic regression of CRC on SNP dosage effect on CRC risk was performed with adjustment for age, sex (when appropriate), centre (when appropriate), smoking status (PHS only), batch effects (ASTERISK only) and the first three PCs from EIGENSTRAT13 to account for population substructure within each individual study. Additional details on sample collection, genotyping, QC and statistical methods are provided in Supplementary Methods.
All samples were collected with informed consent and all procedures were approved by the Human Research Institutional Review Boards (IRBs) at relevant institutions. Specifically, the study protocols of the Japanese and African Americans’ GWASs were approved by the University of Hawaii Human Studies Program and University of Southern California IRB, the IRB in the National Cancer Center, Japan, the Ethics Committee of Kyushu University Faculty of Medical Sciences, the University of North Carolina IRB, Vanderbilt University IRB, the Fred Hutchinson Cancer Research Center IRB and the MD Anderson Cancer Center IRB. The GECCO portion of this work was approved by the Fred Hutchinson Cancer Research Center IRB. The University of Southern California Health Sciences IRB approved all elements of the CORECT study.
Meta-analysis
A fixed-effect model with inverse variance weighting implemented in METAL20 was used to combine the results from the Japanese and the African American studies and for further combining with replication studies. Heterogeneity measure I2 was calculated and Cochran’s Q statistic to test for heterogeneity was calculated21. For the 12 top hits in the VTI1A region at 10q25 (see text), OFCCR in GECCO was excluded because these SNPs did not pass the quality filters in this substudy (Table 1, Supplementary Table 4 and Supplementary Fig. 4). In Supplementary Fig. 5, SNPs that passed the filters in OFCCR were included whenever applicable.
Additional information
How to cite this article: Wang, H. et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat. Commun. 5:4613 doi: 10.1038/ncomms5613 (2014).
References
Lichtenstein, P. et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. New Engl. J. Med. 343, 78–85 (2000).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing data inference for whole genome association studies using localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Wang, H. et al. Fine-mapping of genome-wide association study-identified risk loci for colorectal cancer in African Americans. Hum. Mol. Genet. 22, 5048–5055 (2013).
Bose, A. et al. The v-SNARE Vti1a regulates insulin-stimulated glucose transport and Acrp30 secretion in 3T3-L1 adipocytes. J. Biol. Chem. 280, 36946–36951 (2005).
Kunwar, A. J. et al. Lack of the endosomal SNAREs vti1a and vti1b led to significant impairments in neuronal development. Proc. Natl Acad. Sci. USA 108, 2575–2880 (2011).
Ramirez, D. M., Khvotchev, M., Trauterman, B. & Kavalali, E. T. Vti1a identifies a vesicle pool that preferentially recycles at rest and maintains spontaneous neurotransmission. Neuron 73, 121–134 (2012).
Lan, Q. et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat. Genet. 44, 1330–1335 (2012).
Bass, A. J. et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 43, 964–968 (2011).
The ENCODE Project Consortium. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Loo, L. W. et al. cis-Expression QTL analysis of established colorectal cancer risk variants in colon tumors and adjacent normal tissue. PLoS ONE 7, e30477 (2012).
Cheng, I. et al. Evaluating genetic risk for prostate cancer among Japanese and Latinos. Cancer Epidemiol. Biomarkers Prev. 21, 2048–2058 (2012).
Siddiq, A. et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 21, 5373–5384 (2012).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Sankararaman, S., Sridhar, S., Kimmel, G. & Halperin, E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 9, 290–303 (2008).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Peters, U. et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum. Genet. 131, 217–234 (2012).
Peters, U. et al. Identification of genetic susceptibility loci for colorectal tumors in a genome-wide meta-analysis. Gastroenterology 144, 799–807 (2013).
Zanke, B. W. et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 39, 989–994 (2007).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Higgins, J. P. & Thompson, S. G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Yeager, M. et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649 (2007).
Amundadottir, L. et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 41, 986–990 (2009).
Petersen, G. M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 42, 224–228 (2010).
Landi, M. T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Acknowledgements
We thank Dr Daniel Mirel, who supervised the genotyping of samples while working at the Broad Institute, Boston, MA, and Xin Sheng, Loreall Pooler, Dr Gary K. Chen and Alex H. Stram at the University of Southern California (USC), and Lucy Shen and Mike Loomis at the University of Hawai'i Cancer Center for their technical assistance. The colorectal cancer GWAS among Japanese and African Americans was funded through US National Institutes of Health (NIH) grants 1R01-CA126895, 1R01-CA126895-S1, 1R01-CA104132 and 2U24-CA074806. Genotyping of the additional MEC controls was funded through NIH grants R01-CA132839, RC2-CA148085, R01-CA1326792 and U01-HG004726, as well as a Department of Defense Breast Cancer Research Program, Era of Hope Scholar Award to CAH (W81XWH-08-1-0383). MEC was funded through NIH grants R37 CA54281, P01 CA033619, and R01 CA63464. The SCCS was funded by NIH grant R01CA092447. Data on SCCS cancer cases used in this publication were provided by the Alabama Statewide Cancer Registry; Kentucky Cancer Registry, Lexington, KY; Tennessee Department of Health, Office of Cancer Surveillance; Florida Cancer Data System; North Carolina Central Cancer Registry, North Carolina Division of Public Health; Georgia Comprehensive Cancer Registry; Louisiana Tumor Registry; Mississippi Cancer Registry; South Carolina Central Cancer Registry; Virginia Department of Health, Virginia Cancer Registry; Arkansas Department of Health, Cancer Registry, 4815 W. Markham, Little Rock, AR 72205. The Arkansas Central Cancer Registry is fully funded by a grant from National Program of Cancer Registries, Centers for Disease Control and Prevention (CDC). Data on SCCS cancer cases from Mississippi were collected by the Mississippi Cancer Registry, which participates in the National Program of Cancer Registries (NPCR) of the Centers for Disease Control and Prevention (CDC). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the CDC or the Mississippi Cancer Registry. The UNC studies were supported by grants U01 CA 093326, P50 CA 106991 and R01 CA 66635. The MD Anderson data collection was supported in part by the MD Anderson University Cancer Fund, the MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment, the Center for Clinical and Translational Sciences of the University of Texas Health Science Center at Houston, NCI Cancer Center Support Grant (CA16672) and NCI grant (K07CA160753). JPHC was supported by the National Cancer Center Research and Development Fund (since 2011) and a Grant-in-Aid for Cancer Research (from 1989 to 2010) from the Ministry of Health, Labor and Welfare of Japan. The Fukuoka Colorectal Cancer Study was funded by the Ministry of Education, Culture, Sports, Science and Technology, Japan. The CORECT Study is supported by the National Cancer Institute as part of the GAME-ON consortium (U19 CA148107) with additional support from NCI grants (R01 CA81488, P30 CA014089), the National Human Genome Research Institute at the NIH (T32 HG000040) and the National Institute of Environmental Health Sciences at the NIH (T32 ES013678). CCFR (http://www.coloncfr.org/) is supported by the National Cancer Institute, NIH under RFA #CA-95-011 and through cooperative agreements with members of the Colon Cancer Family Registry and PIs of the Australasian Colorectal Cancer Family Registry (U01 CA097735), Familial Colorectal Neoplasia Collaborative Group (U01 CA074799) [USC], Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783), Seattle Colorectal Cancer Family Registry (U01 CA074794) and the University of Hawaii Colorectal Cancer Family Registry (U01 CA074806). The Colon CFR GWAS work was supported by a National Cancer Institute grant (U01CA122839 and P30 CA014089), Australasian Colorectal Cancer Family Registry (U01 CA097735), Seattle Colorectal Cancer Family Registry (U01 CA074794) and Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute, NIH or any of the collaborating centres in the CCFRs, nor does it mention trade names, commercial products or organizations imply endorsement by the US Government or the CCFR. GECCO was funded by NIH grants U01 CA137088 and R01 CA059045. ASTERISK was supported by a Hospital Clinical Research Program (PHRC) and by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC). CR2&3 was funded by NIH grant R01 CA60987. DACHS was funded by the German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1) and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814). DALS was funded by the NIH (R01 CA48998 to M.L.S.). HPFS is supported by the NIH (P01 CA 055075, UM1 CA167552, R01 137178, and P50 CA 127003), NHS by the NIH (R01 CA137178, P01 CA 087969 and P50 CA 127003) and PHS by the NIH (R01 CA042182). OFCCR was supported by the NIH through funding allocated to the Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); see the CCFR section above. As a subset of ARCTIC, OFCCR is supported by a GL2 grant from the Ontario Research Fund, the Canadian Institutes of Health Research, and the Cancer Risk Evaluation (CaRE) Program grant from the Canadian Cancer Society Research Institute. Thomas J. Hudson and Brent W. Zanke are recipients of Senior Investigator Awards from the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation. PLCO (http://dcp.cancer.gov/plco) was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH. Additionally, a subset of control samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) Prostate Cancer GWAS22, CGEMS pancreatic cancer scan (PanScan)23,24 and the Lung Cancer and Smoking study25. The prostate and PanScan study data sets were accessed with appropriate approval through the dbGaP online resource (http://cgems.cancer.gov/data/) accession numbers phs000207.v1.p1 and phs000206.v3.p2, respectively, and the lung data sets were accessed from the dbGaP website (http://www.ncbi.nlm.nih.gov/gap) through accession number phs000093.v2.p2. Funding for the Lung Cancer and Smoking study was provided by NIH, Genes, Environment and Health Initiative (GEI) Z01 CP 010200, NIH U01 HG004446 and NIH GEI U01 HG 004438. For the lung study, the GENEVA Coordinating Center provided assistance with genotype cleaning and general study coordination, and the Johns Hopkins University Center for Inherited Disease Research conducted genotyping. PMH was funded by the NIH grant R01 CA076366 to P.A. Newcomb. NHS was supported by NIH grants CA 087969, R01 137178, and P50 CA 127003. VITAL was funded by NIH grant K05 CA154337. The WHI program is funded by the National Heart, Lung, and Blood Institute, and NIH through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C and HHSN271201100004C. The ASTERISK study are very grateful to Dr Bruno Buecher, without whom this project would not have existed and also thank all those who agreed to participate in this study, including the patients and the healthy control persons, as well as all the physicians, technicians and students. The DACHS study thank all participants and cooperating clinicians, and Ute Handte-Daub, Renate Hettler-Jensen, Utz Benscheid, Muhabbet Celik and Ursula Eilber at DACHS for excellent technical assistance. GECCO would like to thank all those at the Coordinating Center for helping to bring together the data and people that made this project possible. HPFS, NHS and PHS would like to acknowledge Patrice Soule and Hardeep Ranu of the Dana Farber Harvard Cancer Center High-Throughput Polymorphism Core who assisted in the genotyping for NHS, HPFS, and PHS under the supervision of Dr Immaculata Devivo and Dr David Hunter, Qin (Carolyn) Guo and Lixue Zhu who assisted in programming for NHS and HPFS, and Haiyan Zhang who assisted in programming for the PHS. We would like to thank the participants and staff of the Nurses' Health Study and the Health Professionals Follow-Up Study, for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA and WY. In addition, this study was approved by the Connecticut Department of Public Health (DPH) Human Investigations Committee. Certain data used in this publication were obtained from the DPH. The authors assume full responsibility for analyses and interpretation of these data. The PLCO thank Drs Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff of the PLCO Cancer Screening Trial, Mr. Tom Riley and staff, Information Management Services, Inc., Ms. Barbara O’Brien and staff, Westat, Inc., and Drs Bill Kopp, Wen Shao, and staff, SAIC-Frederick, and most importantly the study participants for their contributions to making this study possible. The PMH study would like to thank the study participants and staff of the Hormones and Colon Cancer study. The WHI study thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: https://cleo.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf.
Author information
Authors and Affiliations
Consortia
Contributions
L.L.M., C.A.H. and D.O.S. contributed to the study concept and design. L.L.M. and T.B. organized the Japanese and African American colorectal cancer consortia. D.V.D.B. supervised genotyping of samples at USC. H.W., L.L.M., D.O.S. and S.L.S. contributed to the statistical analysis. H.W. and L.L.M. drafted the manuscript. L.L.M., L.N.K., B.E.H., S.K., M.I., T.O.K., R.S.S., L.B.S., W.J.B., P.A.N., M.P., C.I.A., D.W.W., S.B., S.I.B., B.W.Z., N.M.L., R.W.H., J.L.H., M.A.J., S.G., G.C., U.P., S.B.G. and S.T. conducted the epidemiological studies that contributed samples to the scan. All authors contributed to the writing of the manuscript, interpretation and discussion of the findings and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
List of members and affiliations appears at the end of the paper
List of members and affiliations appears at the end of the paper
List of members and affiliations appears at the end of the paper
Supplementary information
Supplementary Information
Supplementary Figures 1-5, Supplementary Tables 1-8, Supplementary Methods and Supplementary References (PDF 1299 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Burnett, T., Kono, S. et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat Commun 5, 4613 (2014). https://doi.org/10.1038/ncomms5613
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms5613
This article is cited by
-
Genetic risk impacts the association of menopausal hormone therapy with colorectal cancer risk
British Journal of Cancer (2024)
-
Deciphering colorectal cancer genetics through multi-omic analysis of 100,204 cases and 154,587 controls of European and east Asian ancestries
Nature Genetics (2023)
-
Circulating bilirubin levels and risk of colorectal cancer: serological and Mendelian randomization analyses
BMC Medicine (2020)
-
Diffuse gliomas classified by 1p/19q co-deletion, TERT promoter and IDH mutation status are associated with specific genetic risk loci
Acta Neuropathologica (2018)
-
In silico pathway analysis and tissue specific cis-eQTL for colorectal cancer GWAS risk variants
BMC Genomics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.